Principles of Protein Structure
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protein Structure & Folding
6 Protein Structure & Folding To understand protein folding In the last chapter we learned that proteins are composed of amino acids Goal as a chemical equilibrium. linked together by peptide bonds. We also learned that the twenty amino acids display a wide range of chemical properties. In this chapter we will see Objectives that how a protein folds is determined by its amino acid sequence and that the three-dimensional shape of a folded protein determines its function by After this chapter, you should be able to: the way it positions these amino acids. Finally, we will see that proteins fold • describe the four levels of protein because doing so minimizes Gibbs free energy and that this minimization structure and the thermodynamic involves both making the most favorable bonds and maximizing disorder. forces that stabilize them. • explain how entropy (S) and enthalpy Proteins exhibit four levels of structure (H) contribute to Gibbs free energy. • use the equation ΔG = ΔH – TΔS The structure of proteins can be broken down into four levels of to determine the dependence of organization. The first is primary structure, the linear sequence of amino the favorability of a reaction on acids in the polypeptide chain. By convention, the primary sequence is temperature. written in order from the amino acid at the N-terminus (by convention • explain the hydrophobic effect and its usually on the left) to the amino acid at the C-terminus. The second level role in protein folding. of protein structure, secondary structure, is the local conformation adopted by stretches of contiguous amino acids. -

DNA Glycosylase Exercise - Levels 1 & 2: Answer Key
Name________________________ StarBiochem DNA Glycosylase Exercise - Levels 1 & 2: Answer Key Background In this exercise, you will explore the structure of a DNA repair protein found in most species, including bacteria. DNA repair proteins move along DNA strands, checking for mistakes or damage. DNA glycosylases, a specific type of DNA repair protein, recognize DNA bases that have been chemically altered and remove them, leaving a site in the DNA without a base. Other proteins then come along to fill in the missing DNA base. Learning objectives We will explore the relationship between a protein’s structure and its function in a human DNA glycosylase called human 8-oxoguanine glycosylase (hOGG1). Getting started We will begin this exercise by exploring the structure of hOGG1 using a molecular 3-D viewer called StarBiochem. In this particular structure, the repair protein is bound to a segment of DNA that has been damaged. We will first focus on the structure hOGG1 and then on how this protein interacts with DNA to repair a damaged DNA base. • To begin using StarBiochem, please navigate to: http://mit.edu/star/biochem/. • Click on the Start button Click on the Start button for StarBiochem. • Click Trust when a prompt appears asking if you trust the certificate. • In the top menu, click on Samples à Select from Samples. Within the Amino Acid/Proteins à Protein tab, select “DNA glycosylase hOGG1 w/ DNA – H. sapiens (1EBM)”. “1EBM” is the four character unique ID for this structure. Take a moment to look at the structure from various angles by rotating and zooming on the structure. -

Proteasomes on the Chromosome Cell Research (2017) 27:602-603
602 Cell Research (2017) 27:602-603. © 2017 IBCB, SIBS, CAS All rights reserved 1001-0602/17 $ 32.00 RESEARCH HIGHLIGHT www.nature.com/cr Proteasomes on the chromosome Cell Research (2017) 27:602-603. doi:10.1038/cr.2017.28; published online 7 March 2017 Targeted proteolysis plays an this process, both in mediating homolog ligases, respectively, as RN components important role in the execution and association and in providing crossovers that impact crossover formation [5, 6]. regulation of many cellular events. that tether homologs and ensure their In the first of the two papers, Ahuja et Two recent papers in Science identify accurate segregation. Meiotic recom- al. [7] report that budding yeast pre9∆ novel roles for proteasome-mediated bination is initiated by DNA double- mutants, which lack a nonessential proteolysis in homologous chromo- strand breaks (DSBs) at many sites proteasome subunit, display defects in some pairing, recombination, and along chromosomes, and the multiple meiotic DSB repair, in chromosome segregation during meiosis. interhomolog interactions formed by pairing and synapsis, and in crossover Protein degradation by the 26S pro- DSB repair drive homolog association, formation. Similar defects are seen, teasome drives a variety of processes culminating in the end-to-end homolog but to a lesser extent, in cells treated central to the cell cycle, growth, and dif- synapsis by a protein structure called with the proteasome inhibitor MG132. ferentiation. Proteins are targeted to the the synaptonemal complex (SC) [3]. These defects all can be ascribed to proteasome by covalently ligated chains SC-associated focal protein complexes, a failure to remove nonhomologous of ubiquitin, a small (8.5 kDa) protein. -

Chapter 13 Protein Structure Learning Objectives
Chapter 13 Protein structure Learning objectives Upon completing this material you should be able to: ■ understand the principles of protein primary, secondary, tertiary, and quaternary structure; ■use the NCBI tool CN3D to view a protein structure; ■use the NCBI tool VAST to align two structures; ■explain the role of PDB including its purpose, contents, and tools; ■explain the role of structure annotation databases such as SCOP and CATH; and ■describe approaches to modeling the three-dimensional structure of proteins. Outline Overview of protein structure Principles of protein structure Protein Data Bank Protein structure prediction Intrinsically disordered proteins Protein structure and disease Overview: protein structure The three-dimensional structure of a protein determines its capacity to function. Christian Anfinsen and others denatured ribonuclease, observed rapid refolding, and demonstrated that the primary amino acid sequence determines its three-dimensional structure. We can study protein structure to understand problems such as the consequence of disease-causing mutations; the properties of ligand-binding sites; and the functions of homologs. Outline Overview of protein structure Principles of protein structure Protein Data Bank Protein structure prediction Intrinsically disordered proteins Protein structure and disease Protein primary and secondary structure Results from three secondary structure programs are shown, with their consensus. h: alpha helix; c: random coil; e: extended strand Protein tertiary and quaternary structure Quarternary structure: the four subunits of hemoglobin are shown (with an α 2β2 composition and one beta globin chain high- lighted) as well as four noncovalently attached heme groups. The peptide bond; phi and psi angles The peptide bond; phi and psi angles in DeepView Protein secondary structure Protein secondary structure is determined by the amino acid side chains. -

Introduction to Proteins and Amino Acids Introduction
Introduction to Proteins and Amino Acids Introduction • Twenty percent of the human body is made up of proteins. Proteins are the large, complex molecules that are critical for normal functioning of cells. • They are essential for the structure, function, and regulation of the body’s tissues and organs. • Proteins are made up of smaller units called amino acids, which are building blocks of proteins. They are attached to one another by peptide bonds forming a long chain of proteins. Amino acid structure and its classification • An amino acid contains both a carboxylic group and an amino group. Amino acids that have an amino group bonded directly to the alpha-carbon are referred to as alpha amino acids. • Every alpha amino acid has a carbon atom, called an alpha carbon, Cα ; bonded to a carboxylic acid, –COOH group; an amino, –NH2 group; a hydrogen atom; and an R group that is unique for every amino acid. Classification of amino acids • There are 20 amino acids. Based on the nature of their ‘R’ group, they are classified based on their polarity as: Classification based on essentiality: Essential amino acids are the amino acids which you need through your diet because your body cannot make them. Whereas non essential amino acids are the amino acids which are not an essential part of your diet because they can be synthesized by your body. Essential Non essential Histidine Alanine Isoleucine Arginine Leucine Aspargine Methionine Aspartate Phenyl alanine Cystine Threonine Glutamic acid Tryptophan Glycine Valine Ornithine Proline Serine Tyrosine Peptide bonds • Amino acids are linked together by ‘amide groups’ called peptide bonds. -

Mechanism of Resilin Elasticity
ARTICLE Received 24 Oct 2011 | Accepted 11 Jul 2012 | Published 14 Aug 2012 DOI: 10.1038/ncomms2004 Mechanism of resilin elasticity Guokui Qin1,*, Xiao Hu1,*, Peggy Cebe2 & David L. Kaplan1 Resilin is critical in the flight and jumping systems of insects as a polymeric rubber-like protein with outstanding elasticity. However, insight into the underlying molecular mechanisms responsible for resilin elasticity remains undefined. Here we report the structure and function of resilin from Drosophila CG15920. A reversible beta-turn transition was identified in the peptide encoded by exon III and for full-length resilin during energy input and release, features that correlate to the rapid deformation of resilin during functions in vivo. Micellar structures and nanoporous patterns formed after beta-turn structures were present via changes in either the thermal or the mechanical inputs. A model is proposed to explain the super elasticity and energy conversion mechanisms of resilin, providing important insight into structure–function relationships for this protein. Furthermore, this model offers a view of elastomeric proteins in general where beta-turn-related structures serve as fundamental units of the structure and elasticity. 1 Department of Biomedical Engineering, Tufts University, Medford, Massachusetts 02155, USA. 2 Department of Physics and Astronomy, Tufts University, Medford, Massachusetts 02155, USA. *These authors contributed equally to this work. Correspondence and requests for materials should be addressed to D.L.K. (email: [email protected]). -
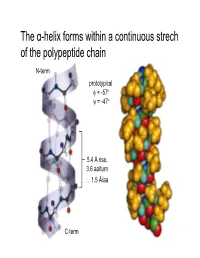
The Α-Helix Forms Within a Continuous Strech of the Polypeptide Chain
The α-helix forms within a continuous strech of the polypeptide chain N-term prototypical φ = -57 ° ψ = -47 ° 5.4 Å rise, 3.6 aa/turn ∴ 1.5 Å/aa C-term α-Helices have a dipole moment, due to unbonded and aligned N-H and C=O groups β-Sheets contain extended (β-strand) segments from separate regions of a protein prototypical φ = -139 °, ψ = +135 ° prototypical φ = -119 °, ψ = +113 ° (6.5Å repeat length in parallel sheet) Antiparallel β-sheets may be formed by closer regions of sequence than parallel Beta turn Figure 6-13 The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups Helix wheel The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups • Occurrence of proline and glycine • Interactions between ends of helix and aa R-groups His Glu N-term C-term -

Structure of the Lifeact–F-Actin Complex
bioRxiv preprint doi: https://doi.org/10.1101/2020.02.16.951269; this version posted February 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Structure of the Lifeact–F-actin complex 2 Alexander Belyy1, Felipe Merino1,2, Oleg Sitsel1 and Stefan Raunser1* 3 1Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Str. 11, 44227 Dortmund, 4 Germany 5 2Current address: Department of Protein Evolution, Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, 6 72076, Tübingen, Germany. 7 *Correspondence should be addressed to: [email protected] 8 9 Abstract 10 Lifeact is a short actin-binding peptide that is used to visualize filamentous actin (F-actin) 11 structures in live eukaryotic cells using fluorescence microscopy. However, this popular probe 12 has been shown to alter cellular morphology by affecting the structure of the cytoskeleton. The 13 molecular basis for such artefacts is poorly understood. Here, we determined the high- 14 resolution structure of the Lifeact–F-actin complex using electron cryo-microscopy. The 15 structure reveals that Lifeact interacts with a hydrophobic binding pocket on F-actin and 16 stretches over two adjacent actin subunits, stabilizing the DNase I-binding loop of actin in the 17 closed conformation. Interestingly, the hydrophobic binding site is also used by actin-binding 18 proteins, such as cofilin and myosin and actin-binding toxins, such as TccC3HVR from 19 Photorhabdus luminescens and ExoY from Pseudomonas aeruginosa. -

Overcharging of Zinc Ion in the Structure of Zinc−Finger Protein Is
Overcharging of zinc ion in the structure of zinc−finger protein is needed for DNA binding stability Ly H. Nguyen,z Tuyen T. Tran,z Truong Thi Ngoc Lien,x Mai Hong Hanh,z and Toan T. Nguyen∗,z zKey Laboratory for Multiscale Simulations of Complex Systems, VNU University of Science, Vietnam National University, Hanoi, 334 Nguyen Trai street, Thanh Xuan district, Hanoi, Vietnam xHanoi University of Science and Technology, 1 Dai Co Viet street, Bach Khoa, Hai Ba Trung district, Hanoi, Vietnam E-mail: [email protected],[email protected] Running header Zinc−finger overcharging arXiv:1911.10452v2 [q-bio.BM] 22 Feb 2020 1 Abstract Zinc finger structure, where a Zn2+ ion binds to four 4 cysteines or histidines in a tetrahedral structure, is a very common motif of nucleic acid−binding proteins. The corresponding interaction model is present in 3% of the genes in human genome. As a result, zinc−finger has been extremely useful in various therapeutic and research capacities, and in biotechnology. In stable configuration of zinc−finger, the cysteine amino acids are deprotonated and become negatively charged. Thus, the Zn2+ ion is overscreened by four cysteine charges (overcharged). Whether this overcharged config- uration is also stable when such a negatively charged zinc−finger binds to a negatively charged DNA molecule is unknown. We investigated how the deprotonated state of cysteine influences its structure, dynamics, and function in binding to DNA molecules by using an all−atom molecular dynamics simulation up to microsecond range of an androgen receptor protein dimer. Our results showed that the deprotonated state of cysteine residues is essential for mechanical stabilization of the functional, folded con- formation. -

The Structure and Function of Large Biological Molecules 5
The Structure and Function of Large Biological Molecules 5 Figure 5.1 Why is the structure of a protein important for its function? KEY CONCEPTS The Molecules of Life Given the rich complexity of life on Earth, it might surprise you that the most 5.1 Macromolecules are polymers, built from monomers important large molecules found in all living things—from bacteria to elephants— can be sorted into just four main classes: carbohydrates, lipids, proteins, and nucleic 5.2 Carbohydrates serve as fuel acids. On the molecular scale, members of three of these classes—carbohydrates, and building material proteins, and nucleic acids—are huge and are therefore called macromolecules. 5.3 Lipids are a diverse group of For example, a protein may consist of thousands of atoms that form a molecular hydrophobic molecules colossus with a mass well over 100,000 daltons. Considering the size and complexity 5.4 Proteins include a diversity of of macromolecules, it is noteworthy that biochemists have determined the detailed structures, resulting in a wide structure of so many of them. The image in Figure 5.1 is a molecular model of a range of functions protein called alcohol dehydrogenase, which breaks down alcohol in the body. 5.5 Nucleic acids store, transmit, The architecture of a large biological molecule plays an essential role in its and help express hereditary function. Like water and simple organic molecules, large biological molecules information exhibit unique emergent properties arising from the orderly arrangement of their 5.6 Genomics and proteomics have atoms. In this chapter, we’ll first consider how macromolecules are built. -

Zinc Finger Proteins: Getting a Grip On
Zinc finger proteins: getting a grip on RNA Raymond S Brown C2H2 (Cys-Cys-His-His motif) zinc finger proteins are members others, such as hZFP100 (C2H2) [11] and tristetraprolin of a large superfamily of nucleic-acid-binding proteins in TTP (CCCH) [12], are involved in histone pre-mRNA eukaryotes. On the basis of NMR and X-ray structures, we processing and the degradation of tumor necrosis factor know that DNA sequence recognition involves a short a helix a mRNA, respectively. In addition, there are reports of bound to the major groove. Exactly how some zinc finger dual RNA/DNA-binding proteins, such as the thyroid proteins bind to double-stranded RNA has been a complete hormone receptor (CCCC) [13] and the trypanosome mystery for over two decades. This has been resolved by the poly-zinc finger PZFP1 pre-mRNA processing protein long-awaited crystal structure of part of the TFIIIA–5S RNA (CCHC) [14]. Whether their interactions with RNA are complex. A comparison can be made with identical fingers in a based on the same mechanisms as protein–DNA binding TFIIIA–DNA structure. Additionally, the NMR structure of is an intriguing structural question that has remained TIS11d bound to an AU-rich element reveals the molecular unanswered until now. details of the interaction between CCCH fingers and single-stranded RNA. Together, these results contrast the What follows is an attempt to expose both similarities different ways that zinc finger proteins bind with high and differences between C2H2 zinc finger protein bind- specificity to their RNA targets. ing to RNA and DNA based on recent X-ray structures. -

Introduction to Protein Structure Prediction
Introduction to Protein Structure Prediction BMI/CS 776 www.biostat.wisc.edu/bmi776/ Colin Dewey [email protected] Spring 2012 The Protein Folding Problem • we know that the function of a protein is determined in large part by its 3D shape (fold, conformation) • can we predict the 3D shape of a protein given only its amino-acid sequence? Protein Architecture • proteins are polymers consisting of amino acids linked by peptide bonds • each amino acid consists of – a central carbon atom (alpha-carbon) – an amino group, NH2 – a carboxyl group, COOH – a side chain • differences in side chains distinguish different amino acids Amino Acids and Peptide Bonds amino side carboxyl group chain group α carbon (common reference point for coordinates of a structure) Amino Acid Side Chains side chains vary in – shape – size – charge – polarity What Determines Conformation? • in general, the amino-acid sequence of a protein determines the 3D shape of a protein [Anfinsen et al., 1950s] • but some qualifications – all proteins can be denatured – some proteins are inherently disordered (i.e. lack a regular structure) – some proteins get folding help from chaperones – there are various mechanisms through which the conformation of a protein can be changed in vivo – post-translational modifications such as phosphorylation – prions – etc. What Determines Conformation? • Which physical properties of the protein determine its fold? – rigidity of the protein backbone – interactions among amino acids, including • electrostatic interactions • van der Waals