A Small Non-Coding RNA Facilitates Brucella Melitensis Intracellular Survival by Regulating the Expression of Virulence Factor T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genital Brucella Suis Biovar 2 Infection of Wild Boar (Sus Scrofa) Hunted in Tuscany (Italy)
microorganisms Article Genital Brucella suis Biovar 2 Infection of Wild Boar (Sus scrofa) Hunted in Tuscany (Italy) Giovanni Cilia * , Filippo Fratini , Barbara Turchi, Marta Angelini, Domenico Cerri and Fabrizio Bertelloni Department of Veterinary Science, University of Pisa, Viale delle Piagge 2, 56124 Pisa, Italy; fi[email protected] (F.F.); [email protected] (B.T.); [email protected] (M.A.); [email protected] (D.C.); [email protected] (F.B.) * Correspondence: [email protected] Abstract: Brucellosis is a zoonosis caused by different Brucella species. Wild boar (Sus scrofa) could be infected by some species and represents an important reservoir, especially for B. suis biovar 2. This study aimed to investigate the prevalence of Brucella spp. by serological and molecular assays in wild boar hunted in Tuscany (Italy) during two hunting seasons. From 287 animals, sera, lymph nodes, livers, spleens, and reproductive system organs were collected. Within sera, 16 (5.74%) were positive to both rose bengal test (RBT) and complement fixation test (CFT), with titres ranging from 1:4 to 1:16 (corresponding to 20 and 80 ICFTU/mL, respectively). Brucella spp. DNA was detected in four lymph nodes (1.40%), five epididymides (1.74%), and one fetus pool (2.22%). All positive PCR samples belonged to Brucella suis biovar 2. The results of this investigation confirmed that wild boar represents a host for B. suis biovar. 2 and plays an important role in the epidemiology of brucellosis in central Italy. Additionally, epididymis localization confirms the possible venereal transmission. Citation: Cilia, G.; Fratini, F.; Turchi, B.; Angelini, M.; Cerri, D.; Bertelloni, Keywords: Brucella suis biovar 2; wild boar; surveillance; epidemiology; reproductive system F. -
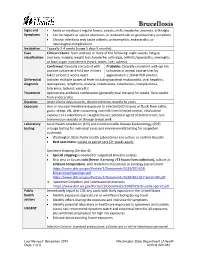
Brucellosis Reporting and Investigation Guideline
Brucellosis Signs and • Acute or insidious irregular fevers, sweats, chills, headache, anorexia, arthralgia Symptoms • Can be hepatic or splenic abscesses, or osteoarticular or genitourinary symptoms • Chronic infections may cause arthritis, osteomyelitis, endocarditis, or neurological complications Incubation Typically 2-4 weeks (range 5 days-5 months) Case Clinical criteria: fever and one or more of the following: night sweats, fatigue, classification anorexia, myalgia, weight loss, headache, arthralgia, arthritis/spondylitis, meningitis, or focal organ involvement (heart, testes, liver, spleen) Confirmed: Clinically consistent with Probable: Clinically consistent with epi link positive culture or 4-fold rise in titers to human or animal case or titer by taken at least 2 weeks apart agglutination ≥ 160 or PCR positive Differential Includes multiple causes of fever including bacterial endocarditis, viral hepatitis, diagnosis leptospirosis, lymphoma, malaria, rickettsioses, tuberculosis, toxoplasmosis, tularemia, typhoid, vasculitis Treatment Appropriate antibiotic combination (generally dual therapy) for weeks. Rare deaths from endocarditis. Duration Acute illness days to week, chronic infection months to years Exposure Skin or mucosal membrane exposure to infected birth tissues or fluids from cattle, goats, sheep, elk, deer; consuming raw milk from infected animal; inhalational exposure in a laboratory or slaughterhouse; potential agent of bioterrorism; rare transmission sexually or through breast milk Laboratory Local Health Jurisdiction -

Brucellosis Annual Report 2018
Brucellosis Annual Report 2018 Brucellosis Brucellosis is a Class A Disease. It must be reported to the state within 24 hours by calling the number listed on the website. Brucellosis is a zoonotic infection of domesticated and wild animals, caused by bacteria of the genus Brucella. Humans become infected by ingestion of food products of animal origin (such as undercooked meat or unpasteurized milk or dairy products), direct contact with infected animals, or inhalation of infectious aerosols. Brucella abortus (cattle), B.melitensis (sheep and goats), B.suis (pigs), and B.canis (dogs), are the most common species. The most common etiology in the U.S. is B.melitensis. Marine Brucella (B.ceti and B.pinnipedialis) may also pose a risk to humans who interact with marine animals; people should avoid contact with stranded or dead marine mammals. Infection may cause a range of symptoms, including fever, sweats, malaise, anorexia, headache, joint and muscle pain, and fatigue. Some symptoms may last for prolonged periods of time including recurrent fevers, arthritis, swelling of the testicle and scrotum area, swelling of the heart, swelling of the liver and/or spleen, neurologic symptoms, chronic fatigue, and depression. Treatment consists of antibiotics, but recovery may take a few weeks to several months. Bovine brucellosis caused by B. abortus, is a bacterial infection transmitted through oral exposure to uterine discharges from infected cows at time of calving or abortion. This previously common disease has been eliminated from the state through the cooperation of the cattle industry and state-federal animal health officials. On November 1, 2000, Louisiana was declared free of brucellosis in cattle. -

Brucellosis History Summary by Russell W
Brucellosis History Summary by Russell W. Currier DVM, MPH Dr. Currier is a DVM, MPH, and Diplomate of the American College for Veterinary Preventive Medicine, who retired from a career as an Iowa State Public Health Veterinarian. Dr. Currier currently presides as President of the American Veterinary Medical History Society. Brucellosis is a sexually transmissible as well as contact-transmitted infection of livestock that can have adverse effects on the fetus and newborn. It occasionally transmits to humans in roles of animal handlers, raw milk consumers, and packing plant workers. In humans, the disease is infectious with long term chronic but distressing febrile episodes rarely fatal with modern clinical management; treatment remains in the domain of infectious disease physicians and should not be attempted by family doctors. The causative organism is Brucella melitensis [from sheep/goats], Brucella bovis [from cattle], Brucella suis [from swine] and Brucella canis [from dogs]. Historically, the disease was recognized in the Mediterranean region, particularly in goats and sheep, dating back to antiquity. Brucellosis-type illnesses were recognized by Hippocrates in his Epidemics writings; the Apostle Paul is considered to have been infected following his being shipwrecked on the Island of Malta and suffered from a recurrent illness or “thorn in my flesh” afterward. Britain maintained a military base on this island during the 18th and 19th centuries. During this period, several British physicians provided vivid descriptions of illness in garrisoned troops and physician David Bruce was dispatched to investigate; he isolated the causative organism from four fatal cases in 1887 and named it Micrococcus melitensis. -

Genome-Resolved Metagenomic Analyses Reveal the Presence of a Putative Bacterial Endosymbiont in an Avian Nasal Mite (Rhinonyssidae; Mesostigmata)
microorganisms Article Genome-Resolved Metagenomic Analyses Reveal the Presence of a Putative Bacterial Endosymbiont in an Avian Nasal Mite (Rhinonyssidae; Mesostigmata) Carolina Osuna-Mascaró 1,*, Jorge Doña 2,3, Kevin P. Johnson 2 and Manuel de Rojas 4,* 1 Department of Biology, University of Nevada, 1664 N Virginia St., Reno, NV 89557, USA 2 Illinois Natural History Survey, Prairie Research Institute, University of Illinois at Urbana-Champaign, Champaign, IL 61820, USA; [email protected] (J.D.); [email protected] (K.P.J.) 3 Departamento de Biología Animal, Universitario de Cartuja, Calle Prof. Vicente Callao, 3, 18011 Granada, Spain 4 Department of Microbiology and Parasitology, Faculty of Pharmacy, Universidad de Sevilla, Calle San Fernando, 4, 41004 Sevilla, Spain * Correspondence: [email protected] (C.O.-M.); [email protected] (M.d.R.) Abstract: Rhinonyssidae (Mesostigmata) is a family of nasal mites only found in birds. All species are hematophagous endoparasites, which may damage the nasal cavities of birds, and also could be potential reservoirs or vectors of other infections. However, the role of members of Rhinonyssidae as disease vectors in wild bird populations remains uninvestigated, with studies of the microbiomes of Rhinonyssidae being almost non-existent. In the nasal mite (Tinaminyssus melloi) from rock doves (Columba livia), a previous study found evidence of a highly abundant putatively endosymbiotic bacteria from Class Alphaproteobacteria. Here, we expanded the sample size of this species (two Citation: Osuna-Mascaró, C.; Doña, different hosts- ten nasal mites from two independent samples per host), incorporated contamination J.; Johnson, K.P.; de Rojas, M. Genome-Resolved Metagenomic controls, and increased sequencing depth in shotgun sequencing and genome-resolved metagenomic Analyses Reveal the Presence of a analyses. -

ESCMID Online Lecture Library © by Author
Arrangement Differentation of anaerobic Classification bacteria according to the Selective media Wadsworth method Level II identification Level III identification of Gram-negative anaerobes Porphyromonas/Prevotella Fusobacterium sp. Level III identification of Gram-positive anaerobes Clostridium sp. Linda Wildeboer-Veloo non-sporeforming rods anaerobic cocci Library Classification (non-) selective media BBA isolation of all anaerobic bacteria Peptostreptococcus Veillonella Brucella Blood Agar Anaerococcus PEA isolation of almost all anaerobic bacteria Peptoniphilus LecturePhenylEthyl Alcohol blood agar Parvimonas Finegoldia BBE isolation of Bacteroides fragilis sp. and Bacteroides Bile Esculin agar Bilophila wadsworthia, inhibition of gal- sensitive bacteria Clostridium-Eubacterium B. fragilis Actinomyces B. ureolyticus BBKV isolation of Bacteroides sp. and Brucella Blood agar with Prevotella sp., inhibition of Gram-positive Propionibacterium Prevotella authorKanamycin and Vancomycin bacteria, promotes pigmentation Bifidobacterium Porphyromonas (laked blood) Lactobacillus-Atopobium Fusobacterium OnlineBilophila by © Additional media Level II indentification CCFA isolation of Clostridium difficile Ka Va Ct bile cat ind Cycloserine Cefoxitin inhibition of almost all anaerobic bacteria Fructose Agar B. fragilis groep RRRRVV Porphyromonas sp. R S R S V +- MMBA isolation of Actinomyces sp. Prevotella sp. RS RVS-V Metronidazole Mupiro- inhibition of most anaerobic bacteria B. ureolyticus SRSS-- ESCMIDcine Blood Agar Bilophila wadsworthia S R S R + - Fusobacterium sp. SRSV-V EYA isolation of lipase- and/or lecithinase positive Veillonella sp. SRSSV- Egg Yolk Agar anaerobic bacteria Ka = kanamycin Va = vancomycin Ct = colistin kat = catalase Wadsworth manual, Table 4-2, p. 58 ind = indole 1 Bacteroides fragilis B. ureolyticus, coloniesLibrary with “pitting” B. wadsworthia on BBA, small translucent colonies. Lecture Gram-stain of B. wadsworthia, straight rods of uniform size and internal vacuoles author Veillonella sp. -

Prepared Culture Media
PREPARED CULTURE MEDIA 030220SG PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modifi ed 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfi eld’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modifi ed 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 -

Historical Perspective of Brucellosis: a Microbiological and Epidemiological Overview
Le Infezioni in Medicina, n. 1, 77-86, 2016 INFECTIONS IN THE HISTORY OF MEDICINE 77 Historical perspective of brucellosis: a microbiological and epidemiological overview Orhan Akpınar Department of Microbiology, Faculty of Dentistry, Suleyman Demirel University, Isparta, Turkey SUMMaRY The historical process of brucellosis extends back to Hippocrates’ time, brucellosis has been characterized by humankind’s first contact with animals. Although bru- fever. Our aim is to investigate selfless work undertaken cellosis is a sporadic disease observed in animals in cer- by scientists on the epidemiology, diagnosis and clinical tain regions of the world, it is an important disease in findings of brucellosis until today, and to gain a histori- humans that can affect many organs and systems due cal perspective about the disease that is as old as human to the consumption of contaminated milk or milk prod- history, still has importance today, causes economic ucts. Studies have shown that the presence of Brucella losses in treated animals and harms human health. dates back to 60 million years ago. In 450 BC, Hippo- crates described a disease similar to brucellosis. Since Keywords: brucellosis, historical perspectives. n INTRODUCTION the abortion risk that may occur due to other in- fectious diseases [2, 3]. The disease is generally rucellosis, which is primarily an animal dis- transmitted to humans by eating cheese, made Bease, is a zoonosis that is transmitted to hu- from raw milk, and milk products such as cream mans from infected animals’ meat, milk, urine, and butter. Brucella can protect their vitality for body fluids, and pregnancy material, as well as 1-2 months in fresh cheese, 4 months in butter, from milk products prepared by infected milk 1 month in ice cream, and 6 months in cream. -

BRUCELLOSIS (Undulant Fever, Malta Fever, Mediterranean Fever, Bang's Disease)
BRUCELLOSIS (Undulant Fever, Malta Fever, Mediterranean Fever, Bang's Disease) REPORTING INFORMATION • Class B: Report by the close of the next business day after the case or suspected case presents and/or a positive laboratory result to the local public health department where the patient resides. If patient residence is unknown, report to the local public health department in which the reporting health care provider or laboratory is located. • Reporting Form(s) and/or Mechanism: o The Ohio Disease Reporting System (ODRS) should be used to report lab findings to the Ohio Department of Health (ODH). For healthcare providers without access to ODRS, you may use the Ohio Confidential Reportable Disease Form (HEA 3334, rev. 1/09 http://www.odh.ohio.gov/wps/portal/gov/odh/know-our-programs/ infectious-disease-control-manual/forms/form-confidential-reportable- disease) • The CDC Brucellosis Case Report Form is required for completion by the local health department. Information collected from the form should be entered into ODRS and faxed to the Ohio Department of Health (ODH) Outbreak Response & Bioterrorism Investigation Team (ORBIT) 614-564-2456. The mailing address for this form is: ODH Outbreak Response & Bioterrorism Investigation Team (ORBIT), 246 N. High St., Columbus, OH 43215. • Key fields for ODRS reporting include: import status (whether the infection was travel-associated or Ohio-acquired), date of illness onset, and all the fields in the AGENTSEpidemiology module. Brucella abortus, Brucella canis, Brucella ceti, Brucella melitensis. Brucella pinnepedialis, and Brucella suis. CASE DEFINITION Clinical Description An illness characterized by acute or insidious onset of fever and one or more of the following: night sweats, arthralgia, headache, fatigue, anorexia, myalgia, weight loss, arthritis/spondylitis, meningitis, or focal organ involvement (endocarditis, orchitis/epididymitis, hepatomegaly, splenomegaly). -

Convergent Evolution of Zoonotic Brucella Species Toward the Selective Use of the Pentose Phosphate Pathway
Convergent evolution of zoonotic Brucella species toward the selective use of the pentose phosphate pathway Arnaud Machelarta,b,1, Kevin Willemarta,1, Amaia Zúñiga-Ripac, Thibault Godardd, Hubert Ploviere, Christoph Wittmannf, Ignacio Moriyónc, Xavier De Bollea,2, Emile Van Schaftingeng,h, Jean-Jacques Letessona,3, and Thibault Barbiera,i,2,3 aResearch Unit in Biology of Microorganisms, Narilis, University of Namur, B-5000 Namur, Belgium; bCenter for Infection and Immunity of Lille, Université de Lille, CNRS, INSERM, Centre Hospitalier Universitaire de Lille, Institut Pasteur de Lille, U1019, Unité Mixtes de Recherche 9017, 59000 Lille, France; cDepartamento de Microbiología e Instituto de Salud Tropical, Instituto de Investigación Sanitaria de Navarra, Universidad de Navarra, 31009 Pamplona, Spain; dInstitute of Biochemical Engineering, Technische Universität Braunschweig, 38106 Braunschweig, Germany; eMetabolism and Nutrition Research Group, Louvain Drug Research Institute, Walloon Excellence in Life Sciences and Biotechnology (WELBIO), Université Catholique de Louvain (UCLouvain), 1200 Brussels, Belgium; fInstitute of Systems Biotechnology, Universität des Saarlandes, 66123 Saarbrücken, Germany; gDe Duve Institute, UCLouvain, 1200 Brussels, Belgium; hWELBIO, UCLouvain, 1200 Brussels, Belgium; and iDepartment of Immunology and Infectious Diseases, Harvard T. H. Chan School of Public Health, Boston, MA 02115 Edited by Roy Curtiss III, University of Florida, Gainesville, FL, and approved August 25, 2020 (received for review May 5, 2020) Mechanistic understanding of the factors that govern host tropism network should similar in all Brucella species (7) and other Rhi- remains incompletely understood for most pathogens. Brucella zobiales (8, 9). It includes all enzymes of the pentose phosphate species, which are capable of infecting a wide range of hosts, offer pathway (PPP), Entner–Doudoroff pathway (EDP), Krebs cycle a useful avenue to address this question. -

An Overview of Brucellosis in Cattle and Humans, and Its Serological and Molecular Diagnosis in Control Strategies
Tropical Medicine and Infectious Disease Review An Overview of Brucellosis in Cattle and Humans, and its Serological and Molecular Diagnosis in Control Strategies Muhammad Zahoor Khan 1 and Muhammad Zahoor 2,* 1 Key Laboratory of Agricultural Animal Genetics and Breeding, National Engineering Laboratory for Animal Breeding, College of Animal Science and Technology, China Agricultural University, Beijing 100193, China; [email protected] 2 Department of Molecular Medicine, Institute of Basic Medical Sciences, University of Oslo, Sognsvannsveien, 90372 Oslo, Norway * Correspondence: [email protected]; Tel.: +47-97-17-8583 Received: 28 April 2018; Accepted: 9 June 2018; Published: 14 June 2018 Abstract: Brucellosis is one of the most common contagious and communicable zoonotic diseases with high rates of morbidity and lifetime sterility. There has been a momentous increase over the recent years in intra/interspecific infection rates, due to poor management and limited resources, especially in developing countries. Abortion in the last trimester is a predominant sign, followed by reduced milk yield and high temperature in cattle, while in humans it is characterized by undulant fever, general malaise, and arthritis. While the clinical picture of brucellosis in humans and cattle is not clear and often misleading with the classical serological diagnosis, efforts have been made to overcome the limitations of current serological assays through the development of PCR-based diagnosis. Due to its complex nature, brucellosis remains a serious threat to public health and livestock in developing countries. In this review, we summarized the recent literature, significant advancements, and challenges in the treatment and vaccination against brucellosis, with a special focus on developing countries. -

Highly Sensitive Bacteriophage-Based Detection of Brucella Abortus in Mixed Culture and Spiked Blood
Article Highly Sensitive Bacteriophage-Based Detection of Brucella abortus in Mixed Culture and Spiked Blood Kirill V. Sergueev, Andrey A. Filippov and Mikeljon P. Nikolich * Department of Bacteriophage Therapeutics, Bacterial Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD 20910, USA; [email protected] (K.V.S.); [email protected] (A.A.F.) * Correspondence: [email protected]; Tel.: +1-301-319-9469 Academic Editor: Tessa E. F. Quax, Matthias G. Fischer and Laurent Debarbieux Received: 1 April 2017; Accepted: 6 June 2017; Published: 10 June 2017 Abstract: For decades, bacteriophages (phages) have been used for Brucella species identification in the diagnosis and epidemiology of brucellosis. Traditional Brucella phage typing is a multi-day procedure including the isolation of a pure culture, a step that can take up to three weeks. In this study, we focused on the use of brucellaphages for sensitive detection of the pathogen in clinical and other complex samples, and developed an indirect method of Brucella detection using real-time quantitative PCR monitoring of brucellaphage DNA amplification via replication on live Brucella cells. This assay allowed the detection of single bacteria (down to 1 colony-forming unit per milliliter) within 72 h without DNA extraction and purification steps. The technique was equally efficient with Brucella abortus pure culture and with mixed cultures of B. abortus and α- proteobacterial near neighbors that can be misidentified as Brucella spp., Ochrobactrum anthropi and Afipia felis. The addition of a simple short sample preparation step enabled the indirect phage-based detection of B.