Sequential Onset and Concurrent Expression of Mir-9 Genomic Loci In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dynamical Gene Regulatory Networks Are Tuned by Transcriptional Autoregulation with Microrna Feedback Thomas G
www.nature.com/scientificreports OPEN Dynamical gene regulatory networks are tuned by transcriptional autoregulation with microRNA feedback Thomas G. Minchington1, Sam Grifths‑Jones2* & Nancy Papalopulu1* Concepts from dynamical systems theory, including multi‑stability, oscillations, robustness and stochasticity, are critical for understanding gene regulation during cell fate decisions, infammation and stem cell heterogeneity. However, the prevalence of the structures within gene networks that drive these dynamical behaviours, such as autoregulation or feedback by microRNAs, is unknown. We integrate transcription factor binding site (TFBS) and microRNA target data to generate a gene interaction network across 28 human tissues. This network was analysed for motifs capable of driving dynamical gene expression, including oscillations. Identifed autoregulatory motifs involve 56% of transcription factors (TFs) studied. TFs that autoregulate have more interactions with microRNAs than non‑autoregulatory genes and 89% of autoregulatory TFs were found in dual feedback motifs with a microRNA. Both autoregulatory and dual feedback motifs were enriched in the network. TFs that autoregulate were highly conserved between tissues. Dual feedback motifs with microRNAs were also conserved between tissues, but less so, and TFs regulate diferent combinations of microRNAs in a tissue‑dependent manner. The study of these motifs highlights ever more genes that have complex regulatory dynamics. These data provide a resource for the identifcation of TFs which regulate the dynamical properties of human gene expression. Cell fate changes are a key feature of development, regeneration and cancer, and are ofen thought of as a “land- scape” that cells move through1,2. Cell fate changes are driven by changes in gene expression: turning genes on or of, or changing their levels above or below a threshold where a cell fate change occurs. -

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -
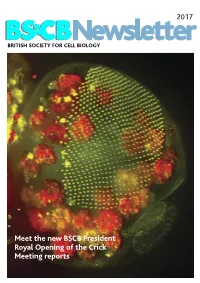
BSCB Newsletter 2017D
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

Analysis of Murine Homeobox Genes Anthony Graham
ANALYSIS OF MURINE HOMEOBOX GENES ANTHONY GRAHAM Thesis for the Degree of Doctor of Philosophy Division of Eukaryotic Molecular Genetics National In s titu te fo r Medical Research The Ridgeway, M ill H ill London NW7 1AA Univeristy College London Gower Street London WC1E 6BT November, 1989 ProQuest Number: 10611102 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a com plete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest ProQuest 10611102 Published by ProQuest LLC(2017). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States C ode Microform Edition © ProQuest LLC. ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106- 1346 This thesis is dedicated to my mother and father. i i The work in this thesis is original except where stated. The sequences used for comparisons in Chapter 3 were not determined by the author but were either from publications from other laboratories or from other members of the laboratory. Anthony Graham: ABSTRACT The work in this thesis is consistent with , and strengthens, suggestions that murine homeobox genes play a role in the establishment of regional specification. It is shown that there is a correlation between the physical order of members of the Hox 2 cluster and their order of expression along the rostrocaudal axis, such that each more 3' gene is expressed more rostrally. -

Stochasticity in the Mir-9/Hes1 Oscillatory Network Can Account For
RESEARCH ARTICLE Stochasticity in the miR-9/Hes1 oscillatory network can account for clonal heterogeneity in the timing of differentiation Nick E Phillips1, Cerys S Manning1†, Tom Pettini1†, Veronica Biga1†, Elli Marinopoulou1†, Peter Stanley1†, James Boyd1, James Bagnall1, Pawel Paszek1, David G Spiller1, Michael RH White1, Marc Goodfellow2,3,4, Tobias Galla5, Magnus Rattray1, Nancy Papalopulu1* 1Faculty of Biology, Medicine and Health, University of Manchester, Manchester, United Kingdom; 2College of Engineering, Mathematics and Physical Sciences, University of Exeter, Exeter, United Kingdom; 3Centre for Biomedical Modelling and Analysis, University of Exeter, Exeter, United Kingdom; 4EPSRC Centre for Predictive Modelling in Healthcare, University of Exeter, Exeter, United Kingdom; 5Theoretical Physics, School of Physics and Astronomy, University of Manchester, Manchester, United Kingdom Abstract Recent studies suggest that cells make stochastic choices with respect to differentiation or division. However, the molecular mechanism underlying such stochasticity is unknown. We previously proposed that the timing of vertebrate neuronal differentiation is *For correspondence: nancy. regulated by molecular oscillations of a transcriptional repressor, HES1, tuned by a post- [email protected] transcriptional repressor, miR-9. Here, we computationally model the effects of intrinsic noise on the Hes1/miR-9 oscillator as a consequence of low molecular numbers of interacting species, †These authors contributed determined experimentally. We report that increased stochasticity spreads the timing of equally to this work differentiation in a population, such that initially equivalent cells differentiate over a period of time. Competing interests: The Surprisingly, inherent stochasticity also increases the robustness of the progenitor state and lessens authors declare that no the impact of unequal, random distribution of molecules at cell division on the temporal spread of competing interests exist. -

MECHANISMS of DEVELOPMENT Now Continued As Cells & Development
MECHANISMS OF DEVELOPMENT Now continued as Cells & Development. AUTHOR INFORMATION PACK TABLE OF CONTENTS XXX . • Description p.1 • Audience p.1 • Impact Factor p.1 • Abstracting and Indexing p.1 • Editorial Board p.2 • Guide for Authors p.3 ISSN: 0925-4773 DESCRIPTION . Mechanisms of Development is now continued as Cells & Development. AUDIENCE . All scientists working in the fields of Cell Biology, Developmental Biology, Molecular Genetics and Differentiation. IMPACT FACTOR . 2019: 2.126 © Clarivate Analytics Journal Citation Reports 2020 ABSTRACTING AND INDEXING . EMBiology Elsevier BIOBASE Biological & Agricultural Index Biological Abstracts BIOSIS Previews Current Awareness in Biological Sciences Genetics Abstracts Science Citation Index BIOSIS Citation Index Chemical Abstracts Current Contents - Life Sciences Embase PubMed/Medline Pascal Francis Scopus AUTHOR INFORMATION PACK 3 Jan 2021 www.elsevier.com/locate/mod 1 EDITORIAL BOARD . Editor-in-Chief Roberto Mayor, University College London, London, United Kingdom Associate Editors Carl-Philipp Heisenberg, Institute of Science and Technology Austria, Klosterneuburg, Austria Sally Horne-Badovinac, University of Chicago, Chicago, Illinois, United States Ana-Maria Lennon-Duménil, Immunity and Cancer, Paris, France Guillaume Salbreux, University of Geneva, Geneva, Switzerland Didier Stainier, Max Planck Institute for Heart and Lung Research, Bad Nauheim, Germany Editorial Board Shinichi Aizawa, RIKEN Center for Developmental Biology, Kobe, Japan Miguel Allende, University of Chile Faculty -

Quantitative Single-Cell Live Imaging Links HES5 Dynamics with Cell-State and Fate in Murine Neurogenesis
ARTICLE https://doi.org/10.1038/s41467-019-10734-8 OPEN Quantitative single-cell live imaging links HES5 dynamics with cell-state and fate in murine neurogenesis Cerys S. Manning 1,7, Veronica Biga1,6, James Boyd 2,6, Jochen Kursawe1,6, Bodvar Ymisson1, David G. Spiller3, Christopher M. Sanderson2, Tobias Galla4, Magnus Rattray 5 & Nancy Papalopulu1,7 1234567890():,; During embryogenesis cells make fate decisions within complex tissue environments. The levels and dynamics of transcription factor expression regulate these decisions. Here, we use single cell live imaging of an endogenous HES5 reporter and absolute protein quantifi- cation to gain a dynamic view of neurogenesis in the embryonic mammalian spinal cord. We report that dividing neural progenitors show both aperiodic and periodic HES5 protein fluctuations. Mathematical modelling suggests that in progenitor cells the HES5 oscillator operates close to its bifurcation boundary where stochastic conversions between dynamics are possible. HES5 expression becomes more frequently periodic as cells transition to dif- ferentiation which, coupled with an overall decline in HES5 expression, creates a transient period of oscillations with higher fold expression change. This increases the decoding capacity of HES5 oscillations and correlates with interneuron versus motor neuron cell fate. Thus, HES5 undergoes complex changes in gene expression dynamics as cells differentiate. 1 School of Medical Sciences, Division of Developmental Biology and Medicine, Faculty of Biology Medicine and Health, The University of Manchester, Oxford Road, Manchester M13 9PT, UK. 2 Department of Cellular and Molecular Physiology, University of Liverpool, Crown Street, Liverpool L69 3BX, UK. 3 School of Biological Sciences, Faculty of Biology Medicine and Health, The University of Manchester, Oxford Road, Manchester M13 9PT, UK. -

Ncomms4399.Pdf
ARTICLE Received 30 Aug 2013 | Accepted 6 Feb 2014 | Published 4 Mar 2014 DOI: 10.1038/ncomms4399 OPEN microRNA input into a neural ultradian oscillator controls emergence and timing of alternative cell states Marc Goodfellow1,w, Nicholas E. Phillips1, Cerys Manning1, Tobias Galla2 & Nancy Papalopulu1 Progenitor maintenance, timed differentiation and the potential to enter quiescence are three fundamental processes that underlie the development of any organ system. In the nervous system, progenitor cells show short-period oscillations in the expression of the transcriptional repressor Hes1, while neurons and quiescent progenitors show stable low and high levels of Hes1, respectively. Here we use experimental data to develop a mathematical model of the double-negative interaction between Hes1 and a microRNA, miR-9, with the aim of understanding how cells transition from one state to another. We show that the input of miR-9 into the Hes1 oscillator tunes its oscillatory dynamics, and endows the system with bistability and the ability to measure time to differentiation. Our results suggest that a relatively simple and widespread network of cross-repressive interactions provides a unifying framework for progenitor maintenance, the timing of differentiation and the emergence of alternative cell states. 1 Faculty of Life Sciences, Michael Smith Building, The University of Manchester, Oxford Road, Manchester M13 9PT, UK. 2 Theoretical Physics, School of Physics and Astronomy, The University of Manchester, Manchester M13 9PL, UK. w Present address: College of Engineering, Mathematics and Physical Sciences, University of Exeter, Exeter, Devon EX4 4QF, UK. Correspondence and requests for materials should be addressed to N.P. (email: [email protected]). -
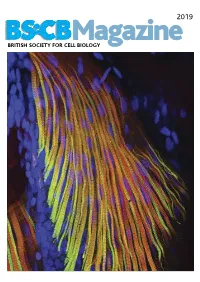
BSCB Newsletter 2019A:BSCB Aut2k7
2019 BSCB Magazine BRITISH SOCIETY FOR CELL BIOLOGY 2019 CONTENTS BSCB Magazine News 2 Book reviews 8 Features 9 Meeting Reports 21 Summer students 25 Society Business 32 Editorial Front cover: microscopic Welcome to the 2019 BSCB Magazine! This year Mustafa Aydogan (University of Oxford), as well as to structure of pectoral fin and Susana and Stephen are filling in for our Newsletter BSCB postdoc poster of the year winners Dr Anna hypaxial muscles of a zebrafish Editor Ann Wheeler. We hope you will enjoy this Caballe (University of Oxford) and Dr Agata Gluszek- Danio rerio larvae at four days year’s magazine! Kustusz (University of Edinburgh). post fertilization. The immunostaining highlights the This year we had a number of fantastic one day In 2019, we will have our jointly BSCB-BSDB organization of fast (red) and meetings sponsored by BSCB. These focus meetings Spring meeting at Warwick University from 7th–10th slow (green) myosins. All nuclei are great way to meet and discuss your science with April, organised by BSCB members Susana Godinho are highlighted in blue (hoechst). experts in your field and to strengthen your network of and Vicky Sanz-Moreno. The programme for this collaborators within the UK. You can read more about meeting, which usually provides a broad spectrum of these meetings in the magazine. If you have an idea themes, has a focus on cancer biology: cell for a focus one day meeting, check how to apply for migration/invasion, organelle biogenesis, trafficking, funding on page 4. Our ambassadors have also been cell-cell communication. -

Expression of Vertebrate Hox Genes During Craniofacial Morphogenesis
Expression of Vertebrate Hox Genes During Craniofacial Morphogenesis Paul Nicholas Hunt Thesis for the Degree of Doctor of Philosophy August 1991 MRC Division of Eukaryotic Molecular Genetics, National Institute of Medical Research, The Ridgeway, Mill Hill, London NW7 1AA University College London, Gower Street, London WC1E 6BT 1 ProQuest Number: 10609329 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a com plete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest ProQuest 10609329 Published by ProQuest LLC(2017). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States C ode Microform Edition © ProQuest LLC. ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106- 1346 Acknowledgements I would like to thank my external supervisor, Dr. Robb Krumlauf, for his excellent help, support and enthusiasm throughout my Ph.D, for many stimulating comments and discussions and for the opportunities for travel to meetings he provided. I want also to thank Dr. Cheryl Tickle for fulfilling the role of internal supervisor and introducing me to craniofacial development. I would like to thank the other members of EKG/K over the years for many interesting discussions and proofreading; Chitrita Chaudhury, Martyn Cook, Heather Marshall, Ian Muchamore, Angela Nieto, Stefan Nonchev, Nancy Papalopulu, Melissa Rubock, and Jenny Whiting. -

Gurdon Institute 2006 PROSPECTUS / ANNUAL REPORT 2005
The Wellcome Trust and Cancer Research UK Gurdon Institute 2006 PROSPECTUS / ANNUAL REPORT 2005 Gurdon I N S T I T U T E PROSPECTUS 2006 ANNUAL REPORT 2005 http://www.gurdon.cam.ac.uk THE GURDON INSTITUTE 1 CONTENTS THE INSTITUTE IN 2005 CHAIRMAN’S INTRODUCTION........................................................3 HISTORICAL BACKGROUND..............................................................4 CENTRAL SUPPORT SERVICES...........................................................5 FUNDING...........................................................................................................5 RESEARCH GROUPS........................................................................6 MEMBERS OF THE INSTITUTE..............................................42 CATEGORIES OF APPOINTMENT..................................................42 POSTGRADUATE OPPORTUNITIES.............................................42 SENIOR GROUP LEADERS..................................................................42 GROUP LEADERS......................................................................................48 SUPPORT STAFF..........................................................................................53 INSTITUTE PUBLICATIONS....................................................55 OTHER INFORMATION STAFF AFFILIATIONS................................................................................62 HONOURS AND AWARDS................................................................63 EDITORIAL BOARDS OF JOURNALS...........................................63 -

Quantitative, Real-Time, Single Cell Analysis in Tissue Reveals Expression Dynamics of Neurogenesis
bioRxiv preprint doi: https://doi.org/10.1101/373407; this version posted July 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Quantitative, real-time, single cell analysis in tissue reveals expression dynamics of neurogenesis Cerys S Manning1*, Veronica Biga1#, James Boyd2#, Jochen Kursawe1#, Bodvar Ymisson1, David G Spiller3, Christopher M Sanderson2, Tobias Galla4, Magnus Rattray5, and Nancy Papalopulu 1* [1] School of Medical Sciences, Division of Developmental Biology and Medicine, Faculty of Biology Medicine and Health, The University of Manchester, Oxford Road, Manchester, M13 9PT, UK [2] Department of Cellular and Molecular Physiology, Crown Street, University of Liverpool, Liverpool, L69 3BX, UK [3] School of Biological Sciences, Faculty of Biology Medicine and Health, The University of Manchester, Oxford Road, Manchester, M13 9PT, UK [4] Theoretical Physics Division, School of Physics and Astronomy, University of Manchester, Manchester, M13 9PL, UK [5] Division of Informatics, Imaging and Data Sciences, Faculty of Biology Medicine and Health, The University of Manchester, Oxford Road, Manchester, M13 9PT, UK * corresponding authors #These authors have contributed equally to this work and are listed alphabetically. Short title: Quantitative and dynamic analysis of neurogenesis bioRxiv preprint doi: https://doi.org/10.1101/373407; this version posted July 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity.