Gurdon Institute 2006 PROSPECTUS / ANNUAL REPORT 2005
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Dynamical Gene Regulatory Networks Are Tuned by Transcriptional Autoregulation with Microrna Feedback Thomas G
www.nature.com/scientificreports OPEN Dynamical gene regulatory networks are tuned by transcriptional autoregulation with microRNA feedback Thomas G. Minchington1, Sam Grifths‑Jones2* & Nancy Papalopulu1* Concepts from dynamical systems theory, including multi‑stability, oscillations, robustness and stochasticity, are critical for understanding gene regulation during cell fate decisions, infammation and stem cell heterogeneity. However, the prevalence of the structures within gene networks that drive these dynamical behaviours, such as autoregulation or feedback by microRNAs, is unknown. We integrate transcription factor binding site (TFBS) and microRNA target data to generate a gene interaction network across 28 human tissues. This network was analysed for motifs capable of driving dynamical gene expression, including oscillations. Identifed autoregulatory motifs involve 56% of transcription factors (TFs) studied. TFs that autoregulate have more interactions with microRNAs than non‑autoregulatory genes and 89% of autoregulatory TFs were found in dual feedback motifs with a microRNA. Both autoregulatory and dual feedback motifs were enriched in the network. TFs that autoregulate were highly conserved between tissues. Dual feedback motifs with microRNAs were also conserved between tissues, but less so, and TFs regulate diferent combinations of microRNAs in a tissue‑dependent manner. The study of these motifs highlights ever more genes that have complex regulatory dynamics. These data provide a resource for the identifcation of TFs which regulate the dynamical properties of human gene expression. Cell fate changes are a key feature of development, regeneration and cancer, and are ofen thought of as a “land- scape” that cells move through1,2. Cell fate changes are driven by changes in gene expression: turning genes on or of, or changing their levels above or below a threshold where a cell fate change occurs. -

Distinctive Regulatory Architectures of Germline-Active and Somatic Genes in C
Downloaded from genome.cshlp.org on October 7, 2021 - Published by Cold Spring Harbor Laboratory Press Research Distinctive regulatory architectures of germline-active and somatic genes in C. elegans Jacques Serizay, Yan Dong, Jürgen Jänes, Michael Chesney, Chiara Cerrato, and Julie Ahringer The Gurdon Institute and Department of Genetics, University of Cambridge, CB2 1QN Cambridge, United Kingdom RNA profiling has provided increasingly detailed knowledge of gene expression patterns, yet the different regulatory ar- chitectures that drive them are not well understood. To address this, we profiled and compared transcriptional and regu- latory element activities across five tissues of Caenorhabditis elegans, covering ∼90% of cells. We find that the majority of promoters and enhancers have tissue-specific accessibility, and we discover regulatory grammars associated with ubiquitous, germline, and somatic tissue–specific gene expression patterns. In addition, we find that germline-active and soma-specific promoters have distinct features. Germline-active promoters have well-positioned +1 and −1 nucleosomes associated with a periodic 10-bp WW signal (W = A/T). Somatic tissue–specific promoters lack positioned nucleosomes and this signal, have wide nucleosome-depleted regions, and are more enriched for core promoter elements, which largely differ between tissues. We observe the 10-bp periodic WW signal at ubiquitous promoters in other animals, suggesting it is an ancient conserved signal. Our results show fundamental differences in regulatory architectures of germline and somatic tissue–specific genes, uncover regulatory rules for generating diverse gene expression patterns, and provide a tissue-specific resource for future studies. [Supplemental material is available for this article.] Cell type–specific transcription regulation underlies production tissues are achieved and whether expression is governed by dis- of the myriad of different cells generated during development. -

Mothers in Science
The aim of this book is to illustrate, graphically, that it is perfectly possible to combine a successful and fulfilling career in research science with motherhood, and that there are no rules about how to do this. On each page you will find a timeline showing on one side, the career path of a research group leader in academic science, and on the other side, important events in her family life. Each contributor has also provided a brief text about their research and about how they have combined their career and family commitments. This project was funded by a Rosalind Franklin Award from the Royal Society 1 Foreword It is well known that women are under-represented in careers in These rules are part of a much wider mythology among scientists of science. In academia, considerable attention has been focused on the both genders at the PhD and post-doctoral stages in their careers. paucity of women at lecturer level, and the even more lamentable The myths bubble up from the combination of two aspects of the state of affairs at more senior levels. The academic career path has academic science environment. First, a quick look at the numbers a long apprenticeship. Typically there is an undergraduate degree, immediately shows that there are far fewer lectureship positions followed by a PhD, then some post-doctoral research contracts and than qualified candidates to fill them. Second, the mentors of early research fellowships, and then finally a more stable lectureship or career researchers are academic scientists who have successfully permanent research leader position, with promotion on up the made the transition to lectureships and beyond. -
BSCB Newsletter 2017D
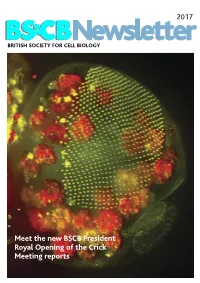
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

President's Message
Winter Issue 2013–2014 SOT News President’s Message I’m starting this President’s message with a quiz!!! It’s just one question, but it’s important that everyone knows the answer. The question is: What do prenatal programming and toxicity, perfluorinalkyl acids ,and human relevance of hemangiosarcomas in rodents have in common? [the answer appears at the end of this message]. While you ponder the answer to that question, I want to reflect on events of this fall and focus on several activities of the Society during recent times of uncertainty. The shutdown of the US government had some effect on nearly all of us. Important meetings, study sections, and day-to- day professional discussion and dialog were all furloughed during this time. However, the most significant impact was on our members who are government employees, and we can only hope that these matters are completely behind us. Unfortunately, there were significant deadlines for SOT matters scheduled during this time, particularly for abstract submissions and award President nominations. Lois D. Lehman- McKeeman I want to specifically acknowledge the work of the Scientific Program and Awards Committees for showing remarkable flexibility in modifying deadlines to accommodate member needs. As a quick review, the Awards committee moved deadlines for nominations to the last possible minute—giving them only about 1 week to review all nominations prior to meeting to select award winners. The prestigious Society awards are central to celebrating member accomplishments, and the work of this committee, against their own time limitations, underscores their commitment to this important activity. -

The ELIXIR Core Data Resources: Fundamental Infrastructure for The
Supplementary Data: The ELIXIR Core Data Resources: fundamental infrastructure for the life sciences The “Supporting Material” referred to within this Supplementary Data can be found in the Supporting.Material.CDR.infrastructure file, DOI: 10.5281/zenodo.2625247 (https://zenodo.org/record/2625247). Figure 1. Scale of the Core Data Resources Table S1. Data from which Figure 1 is derived: Year 2013 2014 2015 2016 2017 Data entries 765881651 997794559 1726529931 1853429002 2715599247 Monthly user/IP addresses 1700660 2109586 2413724 2502617 2867265 FTEs 270 292.65 295.65 289.7 311.2 Figure 1 includes data from the following Core Data Resources: ArrayExpress, BRENDA, CATH, ChEBI, ChEMBL, EGA, ENA, Ensembl, Ensembl Genomes, EuropePMC, HPA, IntAct /MINT , InterPro, PDBe, PRIDE, SILVA, STRING, UniProt ● Note that Ensembl’s compute infrastructure physically relocated in 2016, so “Users/IP address” data are not available for that year. In this case, the 2015 numbers were rolled forward to 2016. ● Note that STRING makes only minor releases in 2014 and 2016, in that the interactions are re-computed, but the number of “Data entries” remains unchanged. The major releases that change the number of “Data entries” happened in 2013 and 2015. So, for “Data entries” , the number for 2013 was rolled forward to 2014, and the number for 2015 was rolled forward to 2016. The ELIXIR Core Data Resources: fundamental infrastructure for the life sciences 1 Figure 2: Usage of Core Data Resources in research The following steps were taken: 1. API calls were run on open access full text articles in Europe PMC to identify articles that mention Core Data Resource by name or include specific data record accession numbers. -

Society for Developmental Biology 64Th Annual Meeting
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Developmental Biology 283 (2005) 537 – 574 www.elsevier.com/locate/ydbio Society for Developmental Biology 64th Annual Meeting Hyatt Regency, San Francisco, CA July 27–August 1, 2005 Organizing Committee: Judith Kimble (Chair, SDB President), Kathy Barton, Minx Fuller, Nipam Patel, Didier Stainier, Xin Sun, Bill Wood Local Organizers: Didier Stainier, Ida Chow Numbers in italics are program abstract numbers and names in bold are speakers. Poster assignments are listed at the end of the meeting program. Program Wednesday, July 27 9 am–5 pm Satellite Symposia (not organized by SDB) Segmentation Seacliff AB Co-organizers: Kenro Kusumi and Olivier Pourquie´ Molecular Biology of Plant Development Seacliff CD Organizer: Kathy Barton 12–7 pm Meeting Registration Grand Foyer Poster Session I and Exhibits Set-up Pacific Concourse See poster assignments at the end of the program 7–9 pm President’s Symposium Grand Ballroom Fundamental Problems of Developmental Biology Chair: Judith Kimble, University of Wisconsin-Madison and HHMI, Madison, WI 7:00 The soma-germline dichotomy. G. Seydoux. Johns Hopkins University, Baltimore, MD 1 7:40 MicroRNAs and their regulatory roles in plants and animals. D.P. Bartel. MIT and Whitehead Institute for Biomedical Research, Cambridge, MA 8:20 The genetics and genomics of evolving new traits in vertebrates. D. Kingsley. Stanford University and HHMI, Stanford, CA 9–11 pm Opening Reception in Honor of Eric Olson, Pacific Concourse Developmental Biology Editor-in-Chief, for invaluable service to the community Poster Session I and Exhibits Pacific Concourse See poster assignments at the end of the program YDBIO-02007; No. -

PLK-1 Promotes the Merger of the Parental Genome Into A
RESEARCH ARTICLE PLK-1 promotes the merger of the parental genome into a single nucleus by triggering lamina disassembly Griselda Velez-Aguilera1, Sylvia Nkombo Nkoula1, Batool Ossareh-Nazari1, Jana Link2, Dimitra Paouneskou2, Lucie Van Hove1, Nicolas Joly1, Nicolas Tavernier1, Jean-Marc Verbavatz3, Verena Jantsch2, Lionel Pintard1* 1Programme Equipe Labe´llise´e Ligue Contre le Cancer - Team Cell Cycle & Development - Universite´ de Paris, CNRS, Institut Jacques Monod, Paris, France; 2Department of Chromosome Biology, Max Perutz Laboratories, University of Vienna, Vienna Biocenter, Vienna, Austria; 3Universite´ de Paris, CNRS, Institut Jacques Monod, Paris, France Abstract Life of sexually reproducing organisms starts with the fusion of the haploid egg and sperm gametes to form the genome of a new diploid organism. Using the newly fertilized Caenorhabditis elegans zygote, we show that the mitotic Polo-like kinase PLK-1 phosphorylates the lamin LMN-1 to promote timely lamina disassembly and subsequent merging of the parental genomes into a single nucleus after mitosis. Expression of non-phosphorylatable versions of LMN- 1, which affect lamina depolymerization during mitosis, is sufficient to prevent the mixing of the parental chromosomes into a single nucleus in daughter cells. Finally, we recapitulate lamina depolymerization by PLK-1 in vitro demonstrating that LMN-1 is a direct PLK-1 target. Our findings indicate that the timely removal of lamin is essential for the merging of parental chromosomes at the beginning of life in C. elegans and possibly also in humans, where a defect in this process might be fatal for embryo development. *For correspondence: [email protected] Introduction Competing interests: The After fertilization, the haploid gametes of the egg and sperm have to come together to form the authors declare that no genome of a new diploid organism. -

Gurdon Institute 20122011 PROSPECTUS / ANNUAL REPORT 20112010
The Wellcome Trust/Cancer Research UK Gurdon Institute 20122011 PROSPECTUS / ANNUAL REPORT 20112010 Gurdon I N S T I T U T E PROSPECTUS 2012 ANNUAL REPORT 2011 http://www.gurdon.cam.ac.uk CONTENTS THE INSTITUTE IN 2011 INTRODUCTION........................................................................................................................................3 HISTORICAL BACKGROUND..........................................................................................................4 CENTRAL SUPPORT SERVICES....................................................................................................5 FUNDING.........................................................................................................................................................5 RETREAT............................................................................................................................................................5 RESEARCH GROUPS.........................................................................................................6 MEMBERS OF THE INSTITUTE................................................................................44 CATEGORIES OF APPOINTMENT..............................................................................44 POSTGRADUATE OPPORTUNITIES..........................................................................44 SENIOR GROUP LEADERS.............................................................................................44 GROUP LEADERS.......................................................................................................................48 -

DECIPHER: Harnessing Local Sequence Context to Improve Protein Multiple Sequence Alignment Erik S
Wright BMC Bioinformatics (2015) 16:322 DOI 10.1186/s12859-015-0749-z RESEARCH ARTICLE Open Access DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment Erik S. Wright1,2 Abstract Background: Alignment of large and diverse sequence sets is a common task in biological investigations, yet there remains considerable room for improvement in alignment quality. Multiple sequence alignment programs tend to reach maximal accuracy when aligning only a few sequences, and then diminish steadily as more sequences are added. This drop in accuracy can be partly attributed to a build-up of error and ambiguity as more sequences are aligned. Most high-throughput sequence alignment algorithms do not use contextual information under the assumption that sites are independent. This study examines the extent to which local sequence context can be exploited to improve the quality of large multiple sequence alignments. Results: Two predictors based on local sequence context were assessed: (i) single sequence secondary structure predictions, and (ii) modulation of gap costs according to the surrounding residues. The results indicate that context-based predictors have appreciable information content that can be utilized to create more accurate alignments. Furthermore, local context becomes more informative as the number of sequences increases, enabling more accurate protein alignments of large empirical benchmarks. These discoveries became the basis for DECIPHER, a new context-aware program for sequence alignment, which outperformed other programs on largesequencesets. Conclusions: Predicting secondary structure based on local sequence context is an efficient means of breaking the independence assumption in alignment. Since secondary structure is more conserved than primary sequence, it can be leveraged to improve the alignment of distantly related proteins. -

Transcriptomic Description of an Endogenous Female State in C
bioRxiv preprint first posted online Oct. 27, 2016; doi: http://dx.doi.org/10.1101/083113. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. It is made available under a CC-BY-NC-ND 4.0 International license. Transcriptomic Description of an Endogenous Female State in C. elegans David Angeles-Albores1,y Daniel H.W. Leighton2,y Tiffany Tsou1 Tiffany H. Khaw1 Igor Antoshechkin3 Paul W. Sternberg1,* October 29, 2016 y These authors contributed equally to this work 1 Department of Biology and Biological Engineering, and Howard Hughes Medical Institute, Caltech, Pasadena, CA, 91125, USA 2 Department of Human Genetics, Department of Biological Chemistry, and Howard Hughes Medical Insti- tute, University of California, Los Angeles, Los Angeles, CA 90095, USA 3 Department of Biology and Biological Engineering, Caltech, Pasadena, CA, 91125, USA * Corresponding author. Contact:[email protected] Abstract Understanding genome and gene function in a whole organism requires us to fully compre- hend the life cycle and the physiology of the organism in question. Although C. elegans is traditionally though of as a hermaphrodite, XX animals exhaust their sperm and become (en- dogenous) females after 3 days of egg-laying. The molecular physiology of this state has not been studied as intensely as other parts of the life cycle, in spite of documented changes in behavior and metabolism that occur at this stage. To study the female state of C. elegans, we designed an experiment to measure the transcriptomes of 1st day adult females; endoge- nous, 6th day adult females; and at the same time points, mutant feminized worms. -

Fish Fins Or Tetrapod Limbs
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector MICHAEL . COATES LIMB EVOLUTION MICHAEL I. COATES LIMB EVOLUTION Fish fins or tetrapod limbs - a simple twist of fate? Comparisons between Hox gene expression patterns in teleost fins and tetrapod limbs are revealing new insights into the developmental mechanisms underlying the evolutionary transition from fin to limb. Phylogenetic trees represent nested patterns of develop- full breadth of the distal mesenchyme, coincident with mental diversification. Yet, until recently, attempts to link development of a 'digital arch' from which the digits will large-scale evolutionary morphological transformations form (see below, Fig 2c) [4]. Thus, HoxD expression to changes in ontogeny have yielded limited results. restriction is reoriented from an antero-posterior to a Explanations have usually characterized developmental proximo-distal axis [1,2]. HoxA expression, in contrast to evolution in terms of shifts in the relative timing of that of the HoxD members, shows no antero-posterior developmental events and alterations to gross embryonic restriction, and instead consists of a series of proximo- patterns, whether static or dynamic, instead of attempting distally nested bands spanning the entire bud (Fig.la). to identify changes affecting specific morphogenetic pro- cesses. But this situation is changing, largely because of The similarities and differences between the development rapid advances in the study of the genes that regulate of the tetrapod limb bud and that of the zebrafish fin bud development. Perhaps most significantly of all, after the are striking (Fig. lb). By comparison with limbs, fin bud initial interest that was inspired by the remarkably con- outgrowth ceases earlier; mesenchymal proliferation fin- served features of some of these developmental genes, ishes as the apical fin bud ectoderm transforms into a pro- research is now beginning to focus on their diversity.