Genomic Analysis of Cichlid Fish 'Natural Mutants'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Impact of Climate Change at Lake Victoria
East Africa Living Lakes Network, C/O OSIENALA (Friends of Lake Victoria). Dunga Beach Kisumu Ann Nabangala Obae. Coordinator. Phone: +254-20-3588681. Email: [email protected] Background It is the Second largest fresh water lake in the World and it is surrounded by three East African States: With 6% in Kenya; Tanzania 52% and Uganda 42%. It is Located at 0:21 0 North and 3:00 0 South of the Equator Lake Victoria has a total length of 3,440 kms and 240 kms wide from East to West and is 1,134 meters above sea level with maximum depth of 82m.Its surface area is 68,870 km 2, catchment area of 180,950 km 2 Generally shallow with maximum depth of 84 meters and mean depth of 40 meters Average inflows and out flows of Lake Victoria Type of flow Flow (m3/s) Percentage (%) Inflows Rain over Lake 3,631 82 Basin Discharge 778 18 Type of flow Flow ( m3/s) Percentage (%) Out flows Evaporation from lake -3,300 76 Nile River - 1,046 24 Balance +33 Sources of Lake Victoria from Kenyan water towers Sondu Miriu river Yala river Nzoia river Mara River Kuja river Impacts and effects Changes in water budget are respectively accompanied by water level fluctuation and promote thermal structures which result in nutrient and food web dynamics. Studies have proved that there is a positive correlation between water level and fish landings (Williams, l972) The abundant fish catches are highly correlated with rainfall and lake levels. The records show that the catches reduced to between 60% and 70% during the current reduction of water level in Nyanza Gulf. -

Evolutionary History of Lake Tanganyika's Predatory Deepwater
Hindawi Publishing Corporation International Journal of Evolutionary Biology Volume 2012, Article ID 716209, 10 pages doi:10.1155/2012/716209 Research Article Evolutionary History of Lake Tanganyika’s Predatory Deepwater Cichlids Paul C. Kirchberger, Kristina M. Sefc, Christian Sturmbauer, and Stephan Koblmuller¨ Department of Zoology, Karl-Franzens-University Graz, Universitatsplatz¨ 2, 8010 Graz, Austria Correspondence should be addressed to Stephan Koblmuller,¨ [email protected] Received 22 December 2011; Accepted 5 March 2012 Academic Editor: R. Craig Albertson Copyright © 2012 Paul C. Kirchberger et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Hybridization among littoral cichlid species in Lake Tanganyika was inferred in several molecular phylogenetic studies. The phenomenon is generally attributed to the lake level-induced shoreline and habitat changes. These allow for allopatric divergence of geographically fragmented populations alternating with locally restricted secondary contact and introgression between incompletely isolated taxa. In contrast, the deepwater habitat is characterized by weak geographic structure and a high potential for gene flow, which may explain the lower species richness of deepwater than littoral lineages. For the same reason, divergent deepwater lineages should have evolved strong intrinsic reproductive isolation already in the incipient stages of diversification, and, consequently, hybridization among established lineages should have been less frequent than in littoral lineages. We test this hypothesis in the endemic Lake Tanganyika deepwater cichlid tribe Bathybatini by comparing phylogenetic trees of Hemibates and Bathybates species obtained with nuclear multilocus AFLP data with a phylogeny based on mitochondrial sequences. -

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

Comparative Analysis of Animal Based Feed Preferences in Selected
International Journal of Fisheries and Aquatic Studies 2019; 7(2): 42-45 E-ISSN: 2347-5129 P-ISSN: 2394-0506 (ICV-Poland) Impact Value: 5.62 Comparative analysis of animal based feed preferences (GIF) Impact Factor: 0.549 IJFAS 2019; 7(2): 42-45 in selected Aquarium fishes © 2019 IJFAS www.fisheriesjournal.com Received: 17-01-2019 Ephsy K Davis and Selvaraju Raja Accepted: 20-02-2019 Ephsy K Davis Abstract Department of Zoology, Ornamental fishes are always an attractive add to your decoration design. In an aquatic ecosystem, the Kongunadu Arts and Science live food organisms constitute the most valuable resources for aquaculture. This outstanding achievement College, Coimbatore, Tamil in animal based feed has resulted in increased survival, higher growth rate and greater resistance to stress. Nadu, India The study of the comparative feeding preferences of ornamental fishes Trichogaster trichopterus(Gourami), Puntius conchonius (Rosybarb), Cyrtocara moorii (Blue Dolphin cichlid), Selvaraju Raja Poecilia sphenops (Blackmolly), Paracheirodon innesi (Neon tetra) towards Mosquito larva, Department of Zoology, Bloodworms and Earthworm has revealed that fishes were fed three feeds and they preferred bloodworm Kongunadu Arts and Science and mosquito larvae. Fish primarily detect food in the aquarium through olfaction (smell) and sight College, Coimbatore, Tamil ratherthan appearance, feel, and taste of the live foods. The Puntius conchonius fish consumed mosquito Nadu, India larvae then earthworm (34.8±7.0 mg and 29.4±4.23 mg) and Trichogaster trichopterus feed more bloodworm (41.25±4.03mg) in short time duration. The selected fishes are indeed suitable feed on mosquito larvae that can be used from wild in the context of mosquito management and inexpensive resources of this larvae can be used for aquarium fish production as alternative potential feed to reduce the feed cost. -

Hohonu Volume 5 (PDF)
HOHONU 2007 VOLUME 5 A JOURNAL OF ACADEMIC WRITING This publication is available in alternate format upon request. TheUniversity of Hawai‘i is an Equal Opportunity Affirmative Action Institution. VOLUME 5 Hohonu 2 0 0 7 Academic Journal University of Hawai‘i at Hilo • Hawai‘i Community College Hohonu is publication funded by University of Hawai‘i at Hilo and Hawai‘i Community College student fees. All production and printing costs are administered by: University of Hawai‘i at Hilo/Hawai‘i Community College Board of Student Publications 200 W. Kawili Street Hilo, Hawai‘i 96720-4091 Phone: (808) 933-8823 Web: www.uhh.hawaii.edu/campuscenter/bosp All rights revert to the witers upon publication. All requests for reproduction and other propositions should be directed to writers. ii d d d d d d d d d d d d d d d d d d d d d d Table of Contents 1............................ A Fish in the Hand is Worth Two on the Net: Don’t Make me Think…different, by Piper Seldon 4..............................................................................................Abortion: Murder-Or Removal of Tissue?, by Dane Inouye 9...............................An Etymology of Four English Words, with Reference to both Grimm’s Law and Verner’s Law by Piper Seldon 11................................Artifacts and Native Burial Rights: Where do We Draw the Line?, by Jacqueline Van Blarcon 14..........................................................................................Ayahuasca: Earth’s Wisdom Revealed, by Jennifer Francisco 16......................................Beak of the Fish: What Cichlid Flocks Reveal About Speciation Processes, by Holly Jessop 26................................................................................. Climatic Effects of the 1815 Eruption of Tambora, by Jacob Smith 33...........................Columnar Joints: An Examination of Features, Formation and Cooling Models, by Mary Mathis 36.................... -

Mayan Cichlid (Cichlasoma Urophthalmum) Ecological Risk Screening Summary
U.S. Fish and Wildlife Service Mayan Cichlid (Cichlasoma urophthalmum) Ecological Risk Screening Summary Web Version – 11/01/2012 Photo: Alexander Calder 1 Native Range, and Status in the United States Native Range From Robins (2001): The Mayan cichlid is native to the Central American Atlantic slope waters of southeastern Mexico (including the Yucatán Peninsula), Belize, Guatemala, Honduras, and Nicaragua. Nonindigenous Occurrences From Schofield et al. (2011): “This species was first documented in Florida when specimens were observed and collected and observed in Everglades National Park in 1983; it is established in several areas in and around the park (Loftus 1987; Lorenz et al. 1997; Smith-Vaniz, personal communication [not cited]; Tilmant 1999) and Big Cypress National Preserve (Nico, unpublished data; Tilmant 1999).” “On the east side of Florida it has been recorded from Canal C-111 north to the Little River Canal (C-7 Canal) (Shafland 1995).” Cichlasoma urophthalmus Ecological Risk Screening Summary U.S. Fish and Wildlife Service – Web Version – 11/1/2012 “A single specimen was taken from a rock pit in Manatee County in October 1975 (Smith- Vaniz, personal communication [not cited]).” “Mayan ciclids have also been collected in Lake Okeechobee and Lake Osbourne, Palm Beach County in 2003 (Cocking 2003; Werner 2003).” “A new population was found in Charlotte Harbor in the summer of 2003 (Adams and Wolfe 2007; Associated Press 2003; Charlotte Harbor NEP 2004; Byrley, personal communication [not cited]). This is the most northern population known.” Reported established in Florida Panther National Wildlife Refuge (2005).” “A specimen was collected in Holiday Park in Broward County (International Game Fishing Association 2000).” “In 2006, this species was found to be established in Mobbly Bayou in Tampa Bay and in canals on Merritt Island in 2007 (Paperno et al. -

Blackchin Tilapia (Sarotherodon Melanotheron) Ecological Risk Screening Summary
U.S. Fish and Wildlife Service Blackchin Tilapia (Sarotherodon melanotheron) Ecological Risk Screening Summary Web Version – 10/01/2012 Photo: © U.S. Geological Survey From Nico and Neilson (2014). 1 Native Range and Nonindigenous Occurrences Native Range From Nico and Neilson (2014): “Tropical Africa. Brackish estuaries and lagoons from Senegal to Zaire (Trewavas 1983).” Nonindigenous Occurrences From Nico and Neilson (2014): “Established in Florida and Hawaii. Evidence indicates it is spreading rapidly in both fresh and salt water around island of Oahu, Hawaii (Devick 1991b).” “The first documented occurrence of this species in Florida was a specimen gillnetted by commercial fishermen in Hillsborough Bay near Tampa, Hillsborough County, in 1959 (Springer and Finucane 1963). Additional records for the western part of the state indicate that this species is established in brackish and freshwaters in eastern Tampa Bay and in adjoining drainages in Hillsborough County, ranging from the Alafia River south to Cockroach Bay. The species has been recorded from the Alafia River from its mouth up to Lithia Springs; from the Hillsborough River, Bullfrog Creek, the Palm River, and the Little Manatee River; and from various western drainage and irrigation ditches (Springer and Finucane 1963; Finucane and Rinckey 1967; Buntz Sarotherodon melanotheron Ecological Risk Screening Summary U.S. Fish and Wildlife Service – Web Version – 10/01/2012 and Manooch 1969; Lachner et al. 1970; Courtenay et al. 1974; Courtenay and Hensley 1979; Courtenay and Kohler 1986; Lee et al. 1980 et seq.; Courtenay and Stauffer 1990; DNR collections; UF museum specimens). There are two records of this species from the west side of Tampa Bay, in Pinellas County: a collection from Lake Maggiore in St. -

SOCIAL ISOLATION and AGGRESSIVENESS in the AMAZONIAN JUVENILE FISH Astronotus Ocellatus
SOCIAL ISOLATION AND AGGRESSIVENESS IN THE AMAZONIAN JUVENILE FISH Astronotus ocellatus GONÇalves-DE-Freitas, E.1 and MARIGUELA, T. C.2 1Laboratório de Comportamento Animal, Departamento de Zoologia e Botânica, IBILCE/UNESP, (CAUNESP, RECAW), R. Cristóvão Colombo, 2265, CEP 15054-000, São José do Rio Preto, SP, Brazil 2Programa de Pós-Graduação em Zoologia, IB, UNESP, Distrito de Rubião Jr, CEP 18618-000, Botucatu, SP, Brazil Correspondence to: Eliane Gonçalves-de-Freitas, Laboratório de Comportamento Animal, Departamento de Zoologia e Botânica, IBILCE/UNESP, (CAUNESP, RECAW), R. Cristóvão Colombo, 2265, CEP 15054-000, São José do Rio Preto, SP, Brazil, e-mail: [email protected] Received July 15, 2003 – Accepted March 23, 2004 – Distributed February 28, 2006 (With 2 figures) ABSTRACT We tested the effect of social isolation on the aggressiveness of an Amazonian fish: Astronotus ocellatus. Ten juvenile fishes were transferred from a group aquarium (60 x 60 x 40 cm) containing 15 individuals (without distinguishing sex) to an isolation aquarium (50 x 40 x 40 cm). Aggressiveness was tested by means of attacks on and displays toward the mirror image. The behavior was video-recorded for 10 min at a time on 4 occasions: at 30 min, 1 day, 5 days and 15 days after isolation. The aggressive drive was analyzed in three ways: latency to display agonistic behavior, frequency of attacks and specific attacks toward the mirror image. The latency to attack decreased during isolation, but the frequency of mouth fighting (a high aggressive attack) tended to increase, indicating an augmented aggressive drive. Our findings are congruent with the behavior of the juvenile cichlid, Haplochromis burtoni but differ from the behavior observed in another cichlid, Pterophylum scalare. -

Molecular Mechanisms Underlying Nuchal Hump Formation in Dolphin Cichlid, Cyrtocara Moorii
www.nature.com/scientificreports OPEN Molecular mechanisms underlying nuchal hump formation in dolphin cichlid, Cyrtocara moorii Laurène Alicia Lecaudey1,2, Christian Sturmbauer1, Pooja Singh1,3 & Ehsan Pashay Ahi1,4* East African cichlid fshes represent a model to tackle adaptive changes and their connection to rapid speciation and ecological distinction. In comparison to bony craniofacial tissues, adaptive morphogenesis of soft tissues has been rarely addressed, particularly at the molecular level. The nuchal hump in cichlids fshes is one such soft-tissue and exaggerated trait that is hypothesized to play an innovative role in the adaptive radiation of cichlids fshes. It has also evolved in parallel across lakes in East Africa and Central America. Using gene expression profling, we identifed and validated a set of genes involved in nuchal hump formation in the Lake Malawi dolphin cichlid, Cyrtocara moorii. In particular, we found genes diferentially expressed in the nuchal hump, which are involved in controlling cell proliferation (btg3, fosl1a and pdgfrb), cell growth (dlk1), craniofacial morphogenesis (dlx5a, mycn and tcf12), as well as regulators of growth-related signals (dpt, pappa and socs2). This is the frst study to identify the set of genes associated with nuchal hump formation in cichlids. Given that the hump is a trait that evolved repeatedly in several African and American cichlid lineages, it would be interesting to see if the molecular pathways and genes triggering hump formation follow a common genetic track or if the trait evolved in parallel, with distinct mechanisms, in other cichlid adaptive radiations and even in other teleost fshes. Given the striking adaptive morphological diversity of craniofacial structures in teleost fsh, it comes with no surprise that these diferences in naturally occurring systems have garnered considerable attention in studies of developmental and molecular biology, beyond models like zebrafsh1,2. -

View/Download
CICHLIFORMES: Cichlidae (part 3) · 1 The ETYFish Project © Christopher Scharpf and Kenneth J. Lazara COMMENTS: v. 6.0 - 30 April 2021 Order CICHLIFORMES (part 3 of 8) Family CICHLIDAE Cichlids (part 3 of 7) Subfamily Pseudocrenilabrinae African Cichlids (Haplochromis through Konia) Haplochromis Hilgendorf 1888 haplo-, simple, proposed as a subgenus of Chromis with unnotched teeth (i.e., flattened and obliquely truncated teeth of H. obliquidens); Chromis, a name dating to Aristotle, possibly derived from chroemo (to neigh), referring to a drum (Sciaenidae) and its ability to make noise, later expanded to embrace cichlids, damselfishes, dottybacks and wrasses (all perch-like fishes once thought to be related), then beginning to be used in the names of African cichlid genera following Chromis (now Oreochromis) mossambicus Peters 1852 Haplochromis acidens Greenwood 1967 acies, sharp edge or point; dens, teeth, referring to its sharp, needle-like teeth Haplochromis adolphifrederici (Boulenger 1914) in honor explorer Adolf Friederich (1873-1969), Duke of Mecklenburg, leader of the Deutsche Zentral-Afrika Expedition (1907-1908), during which type was collected Haplochromis aelocephalus Greenwood 1959 aiolos, shifting, changing, variable; cephalus, head, referring to wide range of variation in head shape Haplochromis aeneocolor Greenwood 1973 aeneus, brazen, referring to “brassy appearance” or coloration of adult males, a possible double entendre (per Erwin Schraml) referring to both “dull bronze” color exhibited by some specimens and to what -

Parte I 70 Genomic Organization and Comparative Chromosome Mapping
!"#$%&'& ()& Genomic organization and comparative chromosome mapping of U1 snRNA gene in cichlid fish, with emphasis in Oreochromis niloticus* D.C. Cabral-de-Mello1,*, G.T. Valente2, R.T. Nakajima2 and C. Martins2 1UNESP – Univ Estadual Paulista, Instituto de Biociências/IB, Departamento de Biologia, Rio Claro, São Paulo, Brazil 2UNESP – Univ Estadual Paulista, Instituto de Biociências/IB, Departamento de Morfologia, Botucatu, São Paulo, Brazil Short running title: Genomic organization and mapping of U1 snRNA in cichlid fish * Corresponding author: UNESP - Univ Estadual Paulista, Instituto de Biociências/IB, Departamento de Biologia, CEP 18618-000 Botucatu, SP, Brazil Phone/Fax: 55 14 38116264. e-mail: [email protected] *Cabral-de-Mello DC, Valente GT, Nakajima RT, Martins C (2012) Genomic organization and comparative chromosome mapping of the U1 snRNA gene in cichlid fish, with an emphasis in Oreochromis niloticus. Chromosome Research 20(2): 279-292. !"#$%&'& (*& Abstract To address the knowledge of genomic and chromosomal organization, and evolutionary patterns of U1 snRNA gene in cichlid fish this gene was cytogenetically mapped and comparatively analyzed in 19 species belonging to several clades of the group. Moreover, the genomic organization of U1 snRNA was analyzed using as reference the Oreochromis niloticus genome. The results indicated a high conservation of one chromosomal cluster of U1 snRNA in the African and Asian species with some level of variation mostly in the South American species. The genomic analysis of U1 revealed a distinct scenario of that observed under the cytogenetic mapping. It was observed just an enrichment of U1 gene in the linkage group (LG) 14, that do not correspond to the same chromosome that harbors the U1 cluster identified under the cytogenetic mapping. -
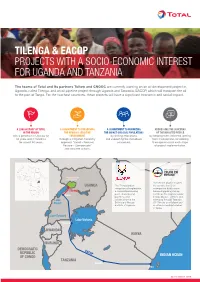
Tilenga & Eacop Projects with a Socio-Economic Interest for Uganda
TILENGA & EACOP PROJECTS WITH A SOCIO-ECONOMIC INTEREST FOR UGANDA AND TANZANIA The teams of Total and its partners Tullow and CNOOC are currently working on an oil development project in Uganda, called Tilenga, and an oil pipeline project through Uganda and Tanzania, EACOP, which will transport the oil to the port of Tanga. For the two host countries, these projects will have a significant economic and social impact. A LONG HISTORY OF TOTAL A COMMITMENT TO PRESERVING A COMMITMENT TO MINIMIZING ADDRESSING THE CONCERNS IN THE REGION THE REGION'S SENSITIVE THE IMPACT ON LOCAL POPULATIONS OF THE IMPACTED PEOPLE with a presence in Uganda for ENVIRONMENT by limiting relocations by keeping them informed, getting 60 years and in Tanzania through a mitigation hierarchy and supporting the individuals them involved and considering for almost 50 years. approach “Avoid – Reduce/ concerned. their opinions into each stage Restore – Compensate” of project implementation. and concrete actions. SOUDAN DU SUD ÉTHIOPIE Murchison Falls National Park The EACOP project involves UGANDA The Tilenga project the construction of an comprises oil exploration, underground hydrocarbon Tilenga a crude oil processing transport pipeline starting Hoima plant, underground just inside the Uganda border pipelines, and (Hoima District - 297km) and Lake infrastructure in the extending through Tanzania Albert Buliisa and Nwoya (1147km) to an oil depot and districts of Uganda. an offshore loading terminal in Tanga. Lake Edward Lake Victoria Bukoba RWANDA KENYA BURUNDI DEMOCRATIC EACOP REPUBLIC INDIAN OCEAN OF CONGO TANZANIA Singida Tanga SEPTEMBER 2019 ZAMBIE MOZAMBIQUE MALAWI SOUDAN DU SUD THIOPIE FOCUS ON THE TILENGA PROJECT Total E&P Uganda, fully aware of the project's sensitive nature, has placed particular emphasis on environmental and societal issues, with a specific commitment to leaving the site in a better state than it was before the work started and to limiting residents' relocations as much as possible.