Root Or Crown: a Developmental Choice Orchestrated by the Differential Regulation of the Epithelial Stem Cell Niche in the Tooth of Two Rodent Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Ono -- PTH-Pthrp Receptor Signaling in Osterix-Expressing Progenitors.Pdf
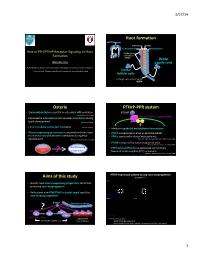
3/27/14 Root forma)on Cementum Dentin Cementoblast Odontoblast Role of PTH/PTHrP Receptor Signaling on Root Epithelial rests Formaon of Malassez (ERM) Dental Wanida Ono papilla cells AGE Orthodon;cs, Department of Developmental Biology, Harvard School of Dental Medicine Endocrine Unit, MassachuseLs General Hospital and Harvard Medical School Dental follicle cells Hertwig’s epithelial root sheath (HERS) Osterix PTHrP-PPR system • Transcripon factor essen;al to osteoblast differen;aon PTHrP (Nakashima K et al. 2002) PTH/PTHrP receptor (PPR) • Expressed in odontoblasts and alveolar osteoblasts during Gαs Gq tooth development (Chen S et al. 2009) • Controls cellular cementum formaon (Cao Z et al. 2012) • Mediates epithelial-mesenchymal interacons • Osterix-expressing precursors in the perichondrium move • PTHrP is expressed in enamel epithelia/HERS? to bone marrow and become osteoblasts during fetal • PPR is expressed in dental mesenchymes development (Maes C, Kronenberg HM et al. 2010) (Beck et al 1995; Lee Deeds and Segre 1995; Liu et al 1998) • PTHrP is required for tooth erup;on in mice (Philbrick WM, Karaplis AC et al. PNAS 1998) Osterix+ Root-forming • PPR haploinsufficiency is associated with primary cells progenitors failure of tooth erup;on (PFE) in humans ? (Decker E, Weber BH et al. Am J Hum Gen 2008) PTHrP expression paern during root morphogenesis Aims of this study PTHrPLacZ/+ x40 P7 P14 P49 • Iden;fy how osterix-expressing progenitors contribute to murine root morphogenesis • Understand how PTH/PTHrP receptor signal regulates root-forming progenitors PTHrP-LacZ x200 P7 x400 P14 ? PTHrP Epithelial root sheath PPR PTHrP-LacZ Osx+ progenitors Odontoblast PTHrP par;cipates in ……. -

Benign Cementoblastoma Associated with an Unerupted Third Molar - a Case Report
CORE Metadata, citation and similar papers at core.ac.uk Provided by Directory of Open Access Journals BENIGN CEMENTOBLASTOMA ASSOCIATED WITH AN UNERUPTED THIRD MOLAR - A CASE REPORT J.Dinakar* M.S.Senthil Kumar** Shiju Mathew Jacob*** *Professor & HOD, Department of Oral Pathology, ** Reader, *** Lecturer, Department of Oral Surgery, Sri Ramakrishna Dental College and Hospital, Coimbatore, Tamilnadu, India. ABSTRACT: Cementoblastoma is a rare odontogenic tumor derived from odontogenic ectomesenchyme of cementoblast origin that forms cementum layer on the roots of a tooth. A case report is presented of a patient treated with surgical excision of Cementoblastoma associated with an unerupted infected right lower third molar tooth. Key words: Cementoblastoma, Odontogenic tumour, unerupted third molar. The cell of origin is cementoblast. INTRODUCTION: Clinically it causes bony expansion. The commonest site is the posterior region of Cementoblastoma is an odontogenic the mandible. In the radiograph it is seen tumor of ectomesenchymal origin. It is as large radiopaque mass associated with also called cementoma. They are large the root of the tooth. We report a case of bulbous mass of cementum or cementum- Benign Cementoblastoma from Sri like tissue on roots of teeth. Ramakrishna Dental College & Hospital, Coimbatore. restriction in opening the mouth and intra oral examination reveals a partially CASE REPORT: erupted third molar tooth with pus discharge. A panoramic radiograph A 41 year old man presented to our showed a radio-opaque, dense, department with a complaint of pain and amorphous, irregularly shaped mass swelling in the right lower half of the face. measuring 2.2 x 1.5cm attached with the Patient gave history of intermittent pain third molar (Fig 1,1a). -

6 Development of the Teeth: Root and Supporting Structures Nagat M
AVERY Chap.06 27-11-2002 10:09 Pagina 108 108 II Development of the Teeth and Supporting Structures 6 Development of the Teeth: Root and Supporting Structures Nagat M. ElNesr and James K. Avery Chapter Outline Introduction Introduction... 108 Objectives... 108 Root development is initiated through the contributions Root Sheath Development... 109 of the cells originating from the enamel organ, dental Single-Root Formation... 110 papilla, and dental follicle. The cells of the outer enamel Multiple-Root Formation... 111 epithelium contact the inner enamel epithelium at the Root Formation Anomalies... 112 base of the enamel organ, the cervical loop (Figs. 6.1 and Fate of the Epithelial Root Sheath (Hertwig's Sheath)... 113 6.2A). Later, with crown completion, the cells of the cer- Dental Follicle... 114 vical loop continue to grow away from the crown and Development of (Intermediate) Cementum... 116 become root sheath cells (Figs. 6.2B and 6.3). The inner Cellular and Acellular Cementum... 116 root sheath cells cause root formation by inducing the Development of the Periodontal Ligament... 117 adjacent cells of the dental papilla to become odonto- Development of the Alveolar Process... 119 blasts, which in turn will form root dentin. The root Summary... 121 sheath will further dictate whether the tooth will have Self-Evaluation Review... 122 single or multiple roots. The remainder of the cells of the dental papilla will then become the cells of the root pulp.The third compo- nent in root formation, the dental follicle, is the tissue that surrounds the enamel organ, the dental papilla, and the root. -

Lecture 2 – Bone
Oral Histology Summary Notes Enoch Ng Lecture 2 – Bone - Protection of brain, lungs, other internal organs - Structural support for heart, lungs, and marrow - Attachment sites for muscles - Mineral reservoir for calcium (99% of body’s) and phosphorous (85% of body’s) - Trap for dangerous minerals (ex:// lead) - Transduction of sound - Endocrine organ (osteocalcin regulates insulin signaling, glucose metabolism, and fat mass) Structure - Compact/Cortical o Diaphysis of long bone, “envelope” of cuboid bones (vertebrae) o 10% porosity, 70-80% calcified (4x mass of trabecular bone) o Protective, subject to bending/torsion/compressive forces o Has Haversian system structure - Trabecular/Cancellous o Metaphysis and epiphysis of long bone, cuboid bone o 3D branching lattice formed along areas of mechanical stress o 50-90% porosity, 15-25% calcified (1/4 mass of compact bone) o High surface area high cellular activity (has marrow) o Metabolic turnover 8x greater than cortical bone o Subject to compressive forces o Trabeculae lined with endosteum (contains osteoprogenitors, osteoblasts, osteoclasts) - Woven Bone o Immature/primitive, rapidly growing . Normally – embryos, newborns, fracture calluses, metaphyseal region of bone . Abnormally – tumors, osteogenesis imperfecta, Pagetic bone o Disorganized, no uniform orientation of collagen fibers, coarse fibers, cells randomly arranged, varying mineral content, isotropic mechanical behavior (behavior the same no matter direction of applied force) - Lamellar Bone o Mature bone, remodeling of woven -

Specialized Stem Cell Niche Enables Repetitive Renewal of Alligator Teeth
Specialized stem cell niche enables repetitive renewal PNAS PLUS of alligator teeth Ping Wua, Xiaoshan Wua,b, Ting-Xin Jianga, Ruth M. Elseyc, Bradley L. Templed, Stephen J. Diverse, Travis C. Glennd, Kuo Yuanf, Min-Huey Cheng,h, Randall B. Widelitza, and Cheng-Ming Chuonga,h,i,1 aDepartment of Pathology, University of Southern California, Los Angeles, CA 90033; bDepartment of Oral and Maxillofacial Surgery, Xiangya Hospital, Central South University, Hunan 410008, China; cLouisiana Department of Wildlife and Fisheries, Rockefeller Wildlife Refuge, Grand Chenier, LA 70643; dEnvironmental Health Science and eDepartment of Small Animal Medicine and Surgery, University of Georgia, Athens, GA 30602; fDepartment of Dentistry and iResearch Center for Wound Repair and Regeneration, National Cheng Kung University, Tainan City 70101, Taiwan; and gSchool of Dentistry and hResearch Center for Developmental Biology and Regenerative Medicine, National Taiwan University, Taipei 10617, Taiwan Edited by Edward M. De Robertis, Howard Hughes Medical Institute/University of California, Los Angeles, CA, and accepted by the Editorial Board March 28, 2013 (received for review July 31, 2012) Reptiles and fish have robust regenerative powers for tooth renewal. replaced from the dental lamina connected to the lingual side of However, extant mammals can either renew their teeth one time the deciduous tooth (15). Human teeth are only replaced one time; (diphyodont dentition) or not at all (monophyodont dentition). however, a remnant of the dental lamina still exists (16) and may Humans replace their milk teeth with permanent teeth and then become activated later in life to form odontogenic tumors (17). lose their ability for tooth renewal. -

Dental Cementum Reviewed: Development, Structure, Composition, Regeneration and Potential Functions
Braz J Oral Sci. January/March 2005 - Vol.4 - Number 12 Dental cementum reviewed: development, structure, composition, regeneration and potential functions Patricia Furtado Gonçalves 1 Enilson Antonio Sallum 1 Abstract Antonio Wilson Sallum 1 This article reviews developmental and structural characteristics of Márcio Zaffalon Casati 1 cementum, a unique avascular mineralized tissue covering the root Sérgio de Toledo 1 surface that forms the interface between root dentin and periodontal Francisco Humberto Nociti Junior 1 ligament. Besides describing the types of cementum and 1 Dept. of Prosthodontics and Periodontics, cementogenesis, attention is given to recent advances in scientific Division of Periodontics, School of Dentistry understanding of the molecular and cellular aspects of the formation at Piracicaba - UNICAMP, Piracicaba, São and regeneration of cementum. The understanding of the mechanisms Paulo, Brazil. involved in the dynamic of this tissue should allow for the development of new treatment strategies concerning the approach of the root surface affected by periodontal disease and periodontal regeneration techniques. Received for publication: October 01, 2004 Key Words: Accepted: December 17, 2004 dental cementum, review Correspondence to: Francisco H. Nociti Jr. Av. Limeira 901 - Caixa Postal: 052 - CEP: 13414-903 - Piracicaba - S.P. - Brazil Tel: ++ 55 19 34125298 Fax: ++ 55 19 3412 5218 E-mail: [email protected] 651 Braz J Oral Sci. 4(12): 651-658 Dental cementum reviewed: development, structure, composition, regeneration and potential functions Introduction junction (Figure 1). The areas and location of acellular Cementum is an avascular mineralized tissue covering the afibrillar cementum vary from tooth to tooth and along the entire root surface. Due to its intermediary position, forming cementoenamel junction of the same tooth6-9. -

Basic Histology (23 Questions): Oral Histology (16 Questions
Board Question Breakdown (Anatomic Sciences section) The Anatomic Sciences portion of part I of the Dental Board exams consists of 100 test items. They are broken up into the following distribution: Gross Anatomy (50 questions): Head - 28 questions broken down in this fashion: - Oral cavity - 6 questions - Extraoral structures - 12 questions - Osteology - 6 questions - TMJ and muscles of mastication - 4 questions Neck - 5 questions Upper Limb - 3 questions Thoracic cavity - 5 questions Abdominopelvic cavity - 2 questions Neuroanatomy (CNS, ANS +) - 7 questions Basic Histology (23 questions): Ultrastructure (cell organelles) - 4 questions Basic tissues - 4 questions Bone, cartilage & joints - 3 questions Lymphatic & circulatory systems - 3 questions Endocrine system - 2 questions Respiratory system - 1 question Gastrointestinal system - 3 questions Genitouirinary systems - (reproductive & urinary) 2 questions Integument - 1 question Oral Histology (16 questions): Tooth & supporting structures - 9 questions Soft oral tissues (including dentin) - 5 questions Temporomandibular joint - 2 questions Developmental Biology (11 questions): Osteogenesis (bone formation) - 2 questions Tooth development, eruption & movement - 4 questions General embryology - 2 questions 2 National Board Part 1: Review questions for histology/oral histology (Answers follow at the end) 1. Normally most of the circulating white blood cells are a. basophilic leukocytes b. monocytes c. lymphocytes d. eosinophilic leukocytes e. neutrophilic leukocytes 2. Blood platelets are products of a. osteoclasts b. basophils c. red blood cells d. plasma cells e. megakaryocytes 3. Bacteria are frequently ingested by a. neutrophilic leukocytes b. basophilic leukocytes c. mast cells d. small lymphocytes e. fibrocytes 4. It is believed that worn out red cells are normally destroyed in the spleen by a. neutrophils b. -

Signaling Networks Regulating Tooth Organogenesis and Regeneration, and the Specification of Dental Mesenchymal and Epithelial Cell Lineages
Downloaded from http://cshperspectives.cshlp.org/ on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press Signaling Networks Regulating Tooth Organogenesis and Regeneration, and the Specification of Dental Mesenchymal and Epithelial Cell Lineages Maria Jussila and Irma Thesleff Developmental Biology Program Institute of Biotechnology, Biokeskus 1, P.O. Box 56, University of Helsinki, Helsinki FIN-00014, Finland Correspondence: maria.jussila@helsinki.fi SUMMARY Teeth develop as ectodermal appendages from epithelial and mesenchymal tissues. Tooth organogenesis is regulated by an intricate network of cell–cell signaling during all steps of development. The dental hard tissues, dentin, enamel, and cementum, are formed by unique cell types whose differentiation is intimately linked with morphogenesis. During evolution the capacity for tooth replacement has been reduced in mammals, whereas teeth have acquired more complex shapes. Mammalian teeth contain stem cells but they may not provide a source for bioengineering of human teeth. Therefore it is likely that nondental cells will have to be reprogrammed for the purpose of clinical tooth regeneration. Obviously this will require understanding of the mechanisms of normal development. The signaling networks mediating the epithelial-mesenchymal interactions during morphogenesis are well characterized but the molecular signatures of the odontogenic tissues remain to be uncovered. Outline 1 Morphogenesis and cell 4 Regulation of tooth replacement, continuous differentiation during tooth development growth, and stem cells in teeth 2 Signal networks and signaling centers 5 Future challenges: stem cell-based bioengineering of teeth 3 Regulation of the identity and 6 Concluding remarks differentiation of odontogenic mesenchymal and epithelial cell lineages References Editors: Patrick P.L. -

Fate Map of the Dental Mesenchyme Dynamic Development Of
Developmental Biology 366 (2012) 244–254 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/locate/developmentalbiology Fate map of the dental mesenchyme: Dynamic development of the dental papilla and follicle Michaela Rothova´ a,b,c, Renata Peterkova´ b, Abigail S. Tucker a,n a Department of Craniofacial Development, King’s College London, Floor 27 Guy’s Tower, Guy’s Hospital, London Bridge, SE1 9RT, London, UK b Department of Teratology, Institute of Experimental Medicine, Academy of Sciences of the Czech Republic, Vı´denskaˇ ´ 1083, 14220 Prague, Czech Republic c Department of Cell Biology, Faculty of Science, Charles University, Vinicˇna´ 7, 128 44 Prague, Czech Republic article info abstract Article history: At the bud stage of tooth development the neural crest derived mesenchyme condenses around the Received 31 October 2011 dental epithelium. As the tooth germ develops and proceeds to the cap stage, the epithelial cervical Received in revised form loops grow and appear to wrap around the condensed mesenchyme, enclosing the cells of the forming 1 March 2012 dental papilla. We have fate mapped the dental mesenchyme, using in vitro tissue culture combined Accepted 30 March 2012 with vital cell labelling and tissue grafting, and show that the dental mesenchyme is a much more Available online 20 April 2012 dynamic population then previously suggested. At the bud stage the mesenchymal cells adjacent to the Keywords: tip of the bud form both the dental papilla and dental follicle. At the early cap stage a small population Tooth of highly proliferative mesenchymal cells in close proximity to the inner dental epithelium and primary Mouse enamel knot provide the major contribution to the dental papilla. -

Sinking Our Teeth in Getting Dental Stem Cells to Clinics for Bone Regeneration
International Journal of Molecular Sciences Review Sinking Our Teeth in Getting Dental Stem Cells to Clinics for Bone Regeneration Sarah Hani Shoushrah , Janis Lisa Transfeld , Christian Horst Tonk, Dominik Büchner , Steffen Witzleben , Martin A. Sieber, Margit Schulze and Edda Tobiasch * Department of Natural Sciences, Bonn-Rhein-Sieg University of Applied Sciences, von-Liebig- Strasse. 20, 53359 Rheinbach, Germany; [email protected] (S.H.S.); [email protected] (J.L.T.); [email protected] (C.H.T.); [email protected] (D.B.); [email protected] (S.W.); [email protected] (M.A.S.); [email protected] (M.S.) * Correspondence: [email protected] Abstract: Dental stem cells have been isolated from the medical waste of various dental tissues. They have been characterized by numerous markers, which are evaluated herein and differentiated into multiple cell types. They can also be used to generate cell lines and iPSCs for long-term in vitro research. Methods for utilizing these stem cells including cellular systems such as organoids or cell sheets, cell-free systems such as exosomes, and scaffold-based approaches with and without drug release concepts are reported in this review and presented with new pictures for clarification. These in vitro applications can be deployed in disease modeling and subsequent pharmaceutical research and also pave the way for tissue regeneration. The main focus herein is on the potential of dental stem cells for hard tissue regeneration, especially bone, by evaluating their potential for osteogenesis Citation: Shoushrah, S.H.; Transfeld, and angiogenesis, and the regulation of these two processes by growth factors and environmental J.L.; Tonk, C.H.; Büchner, D.; stimulators. -

Student to Student Guides
Harvard School of Dental Medicine Student-to-Student Guide to Clinic: How to Excel in Third Year 2010-2011 Edition Adam Donnell Mindy Gil Brandon Grunes Sharon Jin Aram Kim Michelle Mian Tracy Pogal-Sussman Kim Whippy 1999 – Blaine Langberg & Justine Tompkins 2000 – Blaine Langberg & Justine Tompkins 2001 – Blaine Langberg & Justine Tompkins 2002 – Mark Abel & David Halmos 2003 – Ketan Amin 2004 – Rishita Saraiya & Vanessa Yu 2005 – Prathima Prasanna & Amy Crystal 2006 – Seenu Susarla & Brooke Blicher 2007 – Deepak Gupta & Daniel Cassarella 2008 – Bryan Limmer & Josh Kristiansen 2009 – Byran Limmer & Josh Kristiansen 2010 – Adam Donnell, Tracy Pogal-Sussman, Kim Whippy, Mindy Gil, Sharon Jin, Brandon Grunes, Aram Kim, Michelle Mian 1 2 Foreword Dear Class of 2012, We present the 12th edition of this guide to you to assist your transition from the medical school to the HSDM clinic. You have accomplished an enormous amount thus far, but the transformation to come is beyond expectation. Third year is challenging, but fun; you‘ll look back a year from now with amazement at the material you‘ve learned, the skills you‘ve acquired, and the new language that gradually becomes second nature. To ease this process, we would like to share with you the material in this guide, starting with lessons from our own experience. Course material is the bedrock of third year. Without knowing and fully understanding prevention, disease control, and the basics of dentistry, even the most technically skilled dental student can not provide patients with successful treatment. Be on time to lectures, don‘t be afraid to ask questions, and take some time to review your notes in the evening. -

Cell Proliferation Study in Human Tooth Germs
Cell proliferation study in human tooth germs Vanesa Pereira-Prado1, Gabriela Vigil-Bastitta2, Estefania Sicco3, Ronell Bologna-Molina4, Gabriel Tapia-Repetto5 DOI: 10.22592/ode2018n32a10 Abstract The aim of this study was to determine the expression of MCM4-5-6 in human tooth germs in the bell stage. Materials and methods: Histological samples were collected from four fetal maxillae placed in paraffin at the block archive of the Histology Department of the School of Dentistry, UdelaR. Sections were made for HE routine technique and for immunohistochemistry technique for MCM4-5-6. Results: Different regions of the enamel organ showed 100% positivity in the intermediate layer, a variation from 100% to 0% in the inner epithelium from the cervical loop to the incisal area, and 0% in the stellar reticulum as well as the outer epithelium. Conclusions: The results show and confirm the proliferative action of the different areas of the enamel organ. Keywords: MCM4, MCM5, MCM6, tooth germ, cell proliferation. 1 Molecular Pathology in Stomatology, School of Dentistry, Universidad de la República, Montevideo, Uruguay. ORCID: 0000-0001- 7747-671 2 Molecular Pathology in Stomatology, School of Dentistry, Universidad de la República, Montevideo, Uruguay. ORCID: 0000-0002- 0617-1279 3 Molecular Pathology in Stomatology, School of Dentistry, Universidad de la República, Montevideo, Uruguay. ORCID: 0000-0003- 1137-6866 4 Molecular Pathology in Stomatology, School of Dentistry, Universidad de la República, Montevideo, Uruguay. ORCID: 0000-0001- 9755-4779 5 Histology Department, School of Dentistry, Universidad de la República, Montevideo, Uruguay. ORCID: 0000-0003-4563-9142 78 Odontoestomatología. Vol. XX - Nº 32 - Diciembre 2018 Introduction that all the DNA is replicated (12), and prevents DNA from replicating more than once in the Tooth organogenesis is a process involving a same cell cycle (13).