ID Name Description Score P-Value
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
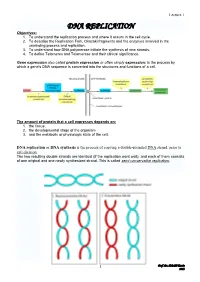
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -
![Downloaded from the NCBI's Genomes Database [123] Or Prot.Pl?Q98JW4 RHILO] and Other Members of Searched Directly Through the NCBI Web Site](https://docslib.b-cdn.net/cover/4253/downloaded-from-the-ncbis-genomes-database-123-or-prot-pl-q98jw4-rhilo-and-other-members-of-searched-directly-through-the-ncbi-web-site-44253.webp)
Downloaded from the NCBI's Genomes Database [123] Or Prot.Pl?Q98JW4 RHILO] and Other Members of Searched Directly Through the NCBI Web Site
BMC Microbiology BioMed Central Research article Open Access A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts Michael Y Galperin* Address: National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA Email: Michael Y Galperin* - [email protected] * Corresponding author Published: 14 June 2005 Received: 18 April 2005 Accepted: 14 June 2005 BMC Microbiology 2005, 5:35 doi:10.1186/1471-2180-5-35 This article is available from: http://www.biomedcentral.com/1471-2180/5/35 © 2005 Galperin; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Analysis of complete microbial genomes showed that intracellular parasites and other microorganisms that inhabit stable ecological niches encode relatively primitive signaling systems, whereas environmental microorganisms typically have sophisticated systems of environmental sensing and signal transduction. Results: This paper presents results of a comprehensive census of signal transduction proteins – histidine kinases, methyl-accepting chemotaxis receptors, Ser/Thr/Tyr protein kinases, adenylate and diguanylate cyclases and c-di-GMP phosphodiesterases – encoded in 167 bacterial and archaeal genomes, sequenced by the end of 2004. The data have been manually checked to avoid false- negative and false-positive hits that commonly arise during large-scale automated analyses and compared against other available resources. The census data show uneven distribution of most signaling proteins among bacterial and archaeal phyla. -

The World of Cyclic Dinucleotides in Bacterial Behavior
molecules Review The World of Cyclic Dinucleotides in Bacterial Behavior Aline Dias da Purificação, Nathalia Marins de Azevedo, Gabriel Guarany de Araujo , Robson Francisco de Souza and Cristiane Rodrigues Guzzo * Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo, São Paulo 01000-000, Brazil * Correspondence: [email protected] or [email protected]; Tel.: +55-11-3091-7298 Received: 27 December 2019; Accepted: 17 March 2020; Published: 25 May 2020 Abstract: The regulation of multiple bacterial phenotypes was found to depend on different cyclic dinucleotides (CDNs) that constitute intracellular signaling second messenger systems. Most notably, c-di-GMP, along with proteins related to its synthesis, sensing, and degradation, was identified as playing a central role in the switching from biofilm to planktonic modes of growth. Recently, this research topic has been under expansion, with the discoveries of new CDNs, novel classes of CDN receptors, and the numerous functions regulated by these molecules. In this review, we comprehensively describe the three main bacterial enzymes involved in the synthesis of c-di-GMP, c-di-AMP, and cGAMP focusing on description of their three-dimensional structures and their structural similarities with other protein families, as well as the essential residues for catalysis. The diversity of CDN receptors is described in detail along with the residues important for the interaction with the ligand. Interestingly, genomic data strongly suggest that there is a tendency for bacterial cells to use both c-di-AMP and c-di-GMP signaling networks simultaneously, raising the question of whether there is crosstalk between different signaling systems. In summary, the large amount of sequence and structural data available allows a broad view of the complexity and the importance of these CDNs in the regulation of different bacterial behaviors. -

Activation-Induced Deoxycytidine Deaminase (AID) Co-Transcriptional Scanning at Single-Molecule Resolution
ARTICLE Received 19 Nov 2014 | Accepted 13 Nov 2015 | Published 18 Dec 2015 DOI: 10.1038/ncomms10209 OPEN Activation-induced deoxycytidine deaminase (AID) co-transcriptional scanning at single-molecule resolution Gayan Senavirathne1,*, Jeffrey G. Bertram2,*, Malgorzata Jaszczur2,*, Kathy R. Chaurasiya3,4,*, Phuong Pham2, Chi H. Mak5,6, Myron F. Goodman2,5 & David Rueda1,3,4 Activation-induced deoxycytidine deaminase (AID) generates antibody diversity in B cells by initiating somatic hypermutation (SHM) and class-switch recombination (CSR) during transcription of immunoglobulin variable (IgV) and switch region (IgS) DNA. Using single- molecule FRET, we show that AID binds to transcribed dsDNA and translocates unidirectionally in concert with RNA polymerase (RNAP) on moving transcription bubbles, while increasing the fraction of stalled bubbles. AID scans randomly when constrained in an 8 nt model bubble. When unconstrained on single-stranded (ss) DNA, AID moves in random bidirectional short slides/hops over the entire molecule while remaining bound for B5 min. Our analysis distinguishes dynamic scanning from static ssDNA creasing. That AID alone can track along with RNAP during transcription and scan within stalled transcription bubbles suggests a mechanism by which AID can initiate SHM and CSR when properly regulated, yet when unregulated can access non-Ig genes and cause cancer. 1 Department of Chemistry, Wayne State University, 5101 Cass Avenue, Detroit, Michigan 48202, USA. 2 Department of Biological Sciences, University of Southern California, Los Angeles, California 90089, USA. 3 Department of Medicine, Section of Virology, Imperial College London, Du Cane Road, London W12 0NN, UK. 4 Single Molecule Imaging Group, MRC Clinical Sciences Center, Imperial College London, Du Cane Road, London W12 0NN, UK. -

The Rnase H-Like Superfamily: New Members, Comparative Structural Analysis and Evolutionary Classification Karolina A
4160–4179 Nucleic Acids Research, 2014, Vol. 42, No. 7 Published online 23 January 2014 doi:10.1093/nar/gkt1414 The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification Karolina A. Majorek1,2,3,y, Stanislaw Dunin-Horkawicz1,y, Kamil Steczkiewicz4, Anna Muszewska4,5, Marcin Nowotny6, Krzysztof Ginalski4 and Janusz M. Bujnicki1,3,* 1Laboratory of Bioinformatics and Protein Engineering, International Institute of Molecular and Cell Biology, ul. Ks. Trojdena 4, PL-02-109 Warsaw, Poland, 2Department of Molecular Physiology and Biological Physics, University of Virginia, 1340 Jefferson Park Avenue, Charlottesville, VA USA-22908, USA, 3Bioinformatics Laboratory, Institute of Molecular Biology and Biotechnology, Adam Mickiewicz University, Umultowska 89, PL-61-614 Poznan, Poland, 4Laboratory of Bioinformatics and Systems Biology, Centre of New Technologies, University of Warsaw, Zwirki i Wigury 93, PL-02-089 Warsaw, Poland, 5Institute of Biochemistry and Biophysics PAS, Pawinskiego 5A, PL-02-106 Warsaw, Poland and 6Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, ul. Ks. Trojdena 4, PL-02-109 Warsaw, Poland Received September 23, 2013; Revised December 12, 2013; Accepted December 26, 2013 ABSTRACT revealed a correlation between the orientation of Ribonuclease H-like (RNHL) superfamily, also called the C-terminal helix with the exonuclease/endo- the retroviral integrase superfamily, groups together nuclease function and the architecture of the numerous enzymes involved in nucleic acid metab- active site. Our analysis provides a comprehensive olism and implicated in many biological processes, picture of sequence-structure-function relation- including replication, homologous recombination, ships in the RNHL superfamily that may guide func- DNA repair, transposition and RNA interference. -

Difluorodeoxycytidine 5'-Triphosphate: a Mechanism of Self-Potentiation1
ICANCER RESEARCH 52, 533-539, February 1, 1992] Cellular Elimination of 2',2'-Difluorodeoxycytidine 5'-Triphosphate: A Mechanism of Self-Potentiation1 Volker Heinemann,2 Y¡-ZhengXu, Sherri Chubb, Alina Sen, Larry W. Hertel, Gerald B. Grindey, and William Plunkett1 Department of Medical Oncology, The University of Texas M. D. Anderson Cancer Center, Houston, Texas 77030 [V. H., Y. X., S. C., A. S., W. P.], and Lilly Research Laboratories, Indianapolis, Indiana 46285 (L. W. H., G. B. G.] ABSTRACT less, clinical cellular pharmacology studies have demonstrated 2',2'-Difluorodeoxycytidine (dFdC, Gemcitabine) is a deoxycytidine that the dFdCTP:dCTP value reaches potentially inhibitory analogue which, after phosphorylation to the 5'-di- and 5'-triphosphate values during clinical trials (4, 10, 11). (b) dFdCTP is incor porated into DNA by DNA polymerases a and f, inhibiting (dFdCTP), induces inhibition of DNA synthesis and cell death. We examined the values for elimination kinetics of cellular dFdCTP and further elongation (9). (c) Once incorporated, dFdCMP resi found they were dependent on cellular concentration after incubation of dues in the terminal or penultimate positions of the DNA strand CCRF-CEM cells with dFdC and washing into drug-free medium. When inhibit the editing function of DNA polymerase <(9). This may the drug was washed out at low cellular dFdCTP levels (<50 n\\), fix damage caused by the incorporated analogue, (d) dFdCDP dFdCTP elimination was linear (t,: = 3.3 h), but it became biphasic at inhibits ribonucleotide reducÃase,blocking DNA synthesis by intracellular dFdCTP levels >100 JIM. Although the initial elimination decreasing the cellular concentrations of deoxynucleoside tri rate was similar at all concentrations, at higher concentrations the phosphates (12-14). -

Phosphatidylinositol-3-Kinase Related Kinases (Pikks) in Radiation-Induced Dna Damage
Mil. Med. Sci. Lett. (Voj. Zdrav. Listy) 2012, vol. 81(4), p. 177-187 ISSN 0372-7025 DOI: 10.31482/mmsl.2012.025 REVIEW ARTICLE PHOSPHATIDYLINOSITOL-3-KINASE RELATED KINASES (PIKKS) IN RADIATION-INDUCED DNA DAMAGE Ales Tichy 1, Kamila Durisova 1, Eva Novotna 1, Lenka Zarybnicka 1, Jirina Vavrova 1, Jaroslav Pejchal 2, Zuzana Sinkorova 1 1 Department of Radiobiology, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic 2 Centrum of Advanced Studies, Faculty of Health Sciences in Hradec Králové, University of Defence in Brno, Czech Republic. Received 5 th September 2012. Revised 27 th November 2012. Published 7 th December 2012. Summary This review describes a drug target for cancer therapy, family of phosphatidylinositol-3 kinase related kinases (PIKKs), and it gives a comprehensive review of recent information. Besides general information about phosphatidylinositol-3 kinase superfamily, it characterizes a DNA-damage response pathway since it is monitored by PIKKs. Key words: PIKKs; ATM; ATR; DNA-PK; Ionising radiation; DNA-repair ABBREVIATIONS therapy and radiation play a pivotal role. Since cancer is one of the leading causes of death worldwide, it is DSB - double stand breaks, reasonable to invest time and resources in the enligh - IR - ionising radiation, tening of mechanisms, which underlie radio-resis - p53 - TP53 tumour suppressors, tance. PI - phosphatidylinositol. The aim of this review is to describe the family INTRODUCTION of phosphatidyinositol 3-kinases (PI3K) and its func - tional subgroup - phosphatidylinositol-3-kinase rela - An efficient cancer treatment means to restore ted kinases (PIKKs) and their relation to repairing of controlled tissue growth via interfering with cell sig - radiation-induced DNA damage. -

The Impact of Genetic Diversity on Gene Essentiality Within the E. Coli Species
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.25.114553; this version posted May 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The impact of genetic diversity on gene essentiality within the E. coli species François Rousset1,2, José Cabezas Caballero1, Florence Piastra-Facon1, Jesús Fernández-Rodríguez3, Olivier Clermont4, Erick Denamur4,5, Eduardo P.C. Rocha6 & David Bikard1,* 1- Synthetic Biology, Department of Microbiology, Institut Pasteur, Paris, France 2- Sorbonne Université, Collège Doctoral, F-75005Paris, France 3- Eligo Bioscience, Paris, France 4- Université de Paris, IAME, INSERM UMR1137, Paris, France 5- AP-HP, Laboratoire de Génétique Moléculaire, Hôpital Bichat, Paris, France 6- Microbial Evolutionary Genomics, Institut Pasteur, CNRS, UMR3525, 25-28 rue Dr Roux, Paris, 75015, France. *To whom correspondence should be addressed: [email protected] Abstract Bacteria from the same species can differ widely in their gene content. In E. coli, the set of genes shared by all strains, known as the core genome, represents about half the number of genes present in any strain. While recent advances in bacterial genomics have enabled to unravel genes required for fitness in various experimental conditions at the genome scale, most studies have focused on model strains. As a result, the impact of this genetic diversity on core processes of the bacterial cell largely remains to be investigated. Here, we developed a new CRISPR interference platform for high- throughput gene repression that is compatible with most E. -

DNA Polymerase V Activity Is Autoregulated by a Novel Intrinsic DNA-Dependent
1 2 DNA polymerase V activity is autoregulated by a novel intrinsic DNA-dependent 3 ATPase 4 Aysen L. Erdem1, Malgorzata Jaszczur1, Jeffrey G. Bertram1, Roger Woodgate2, Michael M. Cox3 & 5 Myron F. Goodman1 6 1Departments of Biological Sciences and Chemistry, University of Southern California, University 7 Park, Los Angeles, California 90089-2910, USA. 2Laboratory of Genomic Integrity, National 8 Institute of Child Health and Human Development, National Institutes of Health, Bethesda, 9 Maryland 20892-3371, USA. 3Department of Biochemistry, University of Wisconsin-Madison, 10 Madison, Wisconsin 53706, USA. 11 12 Escherichia coli DNA polymerase V (pol V), a heterotrimeric complex composed of UmuD′2C, 13 is marginally active. ATP and RecA play essential roles in the activation of pol V for DNA 14 synthesis including translesion synthesis (TLS). We have established three features of the roles 15 of ATP and RecA. 1) RecA-activated DNA polymerase V (pol V Mut), is a DNA-dependent 16 ATPase; 2) bound ATP is required for DNA synthesis; 3) pol V Mut function is regulated by 17 ATP, with ATP required to bind primer/template (p/t) DNA and ATP hydrolysis triggering 18 dissociation from the DNA. Pol V Mut formed with an ATPase-deficient RecA E38K/K72R 19 mutant hydrolyzes ATP rapidly, establishing the DNA-dependent ATPase as an intrinsic 20 property of pol V Mut distinct from the ATP hydrolytic activity of RecA when bound to 21 single-stranded (ss)DNA as a nucleoprotein filament (RecA*). No similar ATPase activity or 22 autoregulatory mechanism has previously been found for a DNA polymerase. -

Effect of Curcumin, Mixture of Curcumin and Piperine and Curcum (Turmeric) on Lipid Profile of Normal and Hyperlipidemic Rats
The Egyptian Journal of Hospital Medicine Vol., 21: 145 – 161 December 2005 I.S.S.N: 12084 2002–1687 Effect of Curcumin, Mixture of Curcumin and Piperine and Curcum (Turmeric) on Lipid Profile of Normal and Hyperlipidemic Rats GHADA, Z. A. Soliman Lecturer of Biochemistry, Biochemistry Department, National Nutrition Institute, Cairo Abstract Curcumin is a polyphenolic, yellow pigment obtained from rhizomes of Curcuma longa (curcum), used as a spice and food colouring. The extracts have several pharmacological effects. We evaluated the effect of curcum, curcumin, and mixture of curcumin and piperine on plasma lipids in normal and hypercholesterolemic rats. A total of 270 rats, divided into 27 groups, were used. G1, G11: control, G2-G11: normal rats fed control diet supplemented with different levels of curcumin and curcum (G2-G6: 0.1%, 0.25%, 0.5%, 1.0%, 2.0% respectively, G7-G11: 1.67%, 4.167%, 8.34%, 16.67%, and 33.34). G12-G26: at first fed control diet supplemented with 2% cholesterol then G13-17, 21-25 fed a control diet supplemented with different levels of curcumin, and curcum [the same levels as G2-G11; G18-20 fed control diet supplemented with mixture of curcumin (0.1, 0.25, 0.5%) and piperine (20 mg/kg BW)], G12 was sacrificed before addition of studied materials, G26 were fed control diet. Lipid profile, triacylglycerol and phospholipids of plasma and organs as liver and heart were measured. Serum cholesterol (total, LDL-C, VLDL-C), triacylglycerol and phospholipids contents were elevated in cholesterol-fed rats, while HDL-C were decreased. -

Mechanistic Study of Physicochemical and Biochemical Processes
Mechanistic study of physicochemical and biochemical processes affecting intestinal absorption of the sesquiterpene lactone nobilin from multi-component systems in the Caco-2 model. Inauguraldissertation zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von URSULA STEPHANIE THORMANN aus Bern (BE) Basel, 2015 Originaldokument gespeichert auf dem Dokumentenserver der Universität Basel edoc.unibas.ch Dieses Werk ist unter dem Vertrag „Creative Commons Namensnennung-Keine kommerzielle Nutzung-Keine Bearbeitung 3.0 Schweiz“ (CC BY-NC-ND 3.0 CH) lizenziert. Die vollständige Lizenz kann unter creativecommons.org/licenses/by-nc-nd/3.0/ch/eingesehen werden. Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. G. Imanidis und Prof. Dr. H. E. U. Meyer zu Schwabedissen Basel, den 18. Februar 2014 Prof. Dr. J. Schibler Namensnennung-Keine kommerzielle Nutzung-Keine Bearbeitung 3.0 Schweiz (CC BY-NC-ND 3.0 CH) Sie dürfen: Teilen — den Inhalt kopieren, verbreiten und zugänglich machen Unter den folgenden Bedingungen: Namensnennung — Sie müssen den Namen des Autors/Rechteinhabers in der von ihm festgelegten Weise nennen. Keine kommerzielle Nutzung — Sie dürfen diesen Inhalt nicht für kommerzielle Zwecke nutzen. Keine Bearbeitung erlaubt — Sie dürfen diesen Inhalt nicht bearbeiten, abwandeln oder in anderer Weise verändern. Wobei gilt: Verzichtserklärung — Jede der vorgenannten Bedingungen kann aufgehoben werden, sofern Sie die