The Dystrophin Node As Integrator of Cytoskeletal Organization, Lateral Force Transmission, Fiber Stability and Cellular Signaling in Skeletal Muscle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Desmin Interacts Directly with Mitochondria
International Journal of Molecular Sciences Article Desmin Interacts Directly with Mitochondria Alexander A. Dayal 1, Natalia V. Medvedeva 1, Tatiana M. Nekrasova 1, Sergey D. Duhalin 1, Alexey K. Surin 1,2 and Alexander A. Minin 1,* 1 Institute of Protein Research of Russian Academy of Sciences, Vavilova st., 34, 119334 Moscow, Russia; [email protected] (A.A.D.); [email protected] (N.V.M.); [email protected] (T.M.N.); [email protected] (S.D.D.); [email protected] (A.K.S.) 2 Pushchino Branch, Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Prospekt Nauki 6, Pushchino, 142290 Moscow Region, Russia * Correspondence: [email protected] Received: 14 October 2020; Accepted: 26 October 2020; Published: 30 October 2020 Abstract: Desmin intermediate filaments (IFs) play an important role in maintaining the structural and functional integrity of muscle cells. They connect contractile myofibrils to plasma membrane, nuclei, and mitochondria. Disturbance of their network due to desmin mutations or deficiency leads to an infringement of myofibril organization and to a deterioration of mitochondrial distribution, morphology, and functions. The nature of the interaction of desmin IFs with mitochondria is not clear. To elucidate the possibility that desmin can directly bind to mitochondria, we have undertaken the study of their interaction in vitro. Using desmin mutant Des(Y122L) that forms unit-length filaments (ULFs) but is incapable of forming long filaments and, therefore, could be effectively separated from mitochondria by centrifugation through sucrose gradient, we probed the interaction of recombinant human desmin with mitochondria isolated from rat liver. Our data show that desmin can directly bind to mitochondria, and this binding depends on its N-terminal domain. -

Supplementary Data
Figure 2S 4 7 A - C 080125 CSCs 080418 CSCs - + IFN-a 48 h + IFN-a 48 h + IFN-a 72 h 6 + IFN-a 72 h 3 5 MRFI 4 2 3 2 1 1 0 0 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 7 B 13 080125 FBS - D 080418 FBS - + IFN-a 48 h 12 + IFN-a 48 h + IFN-a 72 h + IFN-a 72 h 6 080125 FBS 11 10 5 9 8 4 7 6 3 MRFI 5 4 2 3 2 1 1 0 0 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 MHC I MHC II MICA MICB ULBP-1 ULBP-2 ULBP-3 ULBP-4 Molecule Molecule FIGURE 4S FIGURE 5S Panel A Panel B FIGURE 6S A B C D Supplemental Results Table 1S. Modulation by IFN-α of APM in GBM CSC and FBS tumor cell lines. Molecule * Cell line IFN-α‡ HLA β2-m# HLA LMP TAP1 TAP2 class II A A HC§ 2 7 10 080125 CSCs - 1∞ (1) 3 (65) 2 (91) 1 (2) 6 (47) 2 (61) 1 (3) 1 (2) 1 (3) + 2 (81) 11 (80) 13 (99) 1 (3) 8 (88) 4 (91) 1 (2) 1 (3) 2 (68) 080125 FBS - 2 (81) 4 (63) 4 (83) 1 (3) 6 (80) 3 (67) 2 (86) 1 (3) 2 (75) + 2 (99) 14 (90) 7 (97) 5 (75) 7 (100) 6 (98) 2 (90) 1 (4) 3 (87) 080418 CSCs - 2 (51) 1 (1) 1 (3) 2 (47) 2 (83) 2 (54) 1 (4) 1 (2) 1 (3) + 2 (81) 3 (76) 5 (75) 2 (50) 2 (83) 3 (71) 1 (3) 2 (87) 1 (2) 080418 FBS - 1 (3) 3 (70) 2 (88) 1 (4) 3 (87) 2 (76) 1 (3) 1 (3) 1 (2) + 2 (78) 7 (98) 5 (99) 2 (94) 5 (100) 3 (100) 1 (4) 2 (100) 1 (2) 070104 CSCs - 1 (2) 1 (3) 1 (3) 2 (78) 1 (3) 1 (2) 1 (3) 1 (3) 1 (2) + 2 (98) 8 (100) 10 (88) 4 (89) 3 (98) 3 (94) 1 (4) 2 (86) 2 (79) * expression of APM molecules was evaluated by intracellular staining and cytofluorimetric analysis; ‡ cells were treatead or not (+/-) for 72 h with 1000 IU/ml of IFN-α; # β-2 microglobulin; § β-2 microglobulin-free HLA-A heavy chain; ∞ values are indicated as ratio between the mean of fluorescence intensity of cells stained with the selected mAb and that of the negative control; bold values indicate significant MRFI (≥ 2). -

Microanatomy of Muscles
Microanatomy of Muscles Anatomy & Physiology Class Three Main Muscle Types Objectives: By the end of this presentation you will have the information to: 1. Describe the 3 main types of muscles. 2. Detail the functions of the muscle system. 3. Correctly label the parts of a myocyte (muscle cell) 4. Identify the levels of organization in a skeletal muscle from organ to myosin. 5. Explain how a muscle contracts utilizing the correct terminology of the sliding filament theory. 6. Contrast and compare cardiac and smooth muscle with skeletal muscle. Major Functions: Muscle System 1. Moving the skeletal system and posture. 2. Passing food through the digestive system & constriction of other internal organs. 3. Production of body heat. 4. Pumping the blood throughout the body. 5. Communication - writing and verbal Specialized Cells (Myocytes) ~ Contractile Cells Can shorten along one or more planes because of specialized cell membrane (sarcolemma) and specialized cytoskeleton. Specialized Structures found in Myocytes Sarcolemma: The cell membrane of a muscle cell Transverse tubule: a tubular invagination of the sarcolemma of skeletal or cardiac muscle fibers that surrounds myofibrils; involved in transmitting the action potential from the sarcolemma to the interior of the myofibril. Sarcoplasmic Reticulum: The special type of smooth endoplasmic Myofibrils: reticulum found in smooth and a contractile fibril of skeletal muscle, composed striated muscle fibers whose function mainly of actin and myosin is to store and release calcium ions. Multiple Nuclei (skeletal) & many mitochondria Skeletal Muscle - Microscopic Anatomy A whole skeletal muscle (such as the biceps brachii) is considered an organ of the muscular system. Each organ consists of skeletal muscle tissue, connective tissue, nerve tissue, and blood or vascular tissue. -

In Situlocalization of Cytoskeletal Elements in the Human Trabecular
Investigative Ophthalmology & Visual Science. Vol. 31. No. 9. September 1990 Cops right £• Association lor Research in Vision and Ophthalmology In Situ Localization of Cytoskeletal Elements in the Human Trabecular Meshwork and Cornea Robert N. Weinreb* and Mark I. Ryderf The authors compared cytoskeletal elements of the in situ human trabccular-mcshwork cell with in situ human corneal cells using indirect immunofluorcsccncc staining for tubulin and intermediate filaments (vimentin, cytokeratin, and desmin) and NBD-phallacidin staining for f-actin using both fixed frozen and unfixed frozen sections from postmortem eyes. Both f-actin and tubulin were found throughout the cell body of trabecular-meshwork cells, keratocytes, corneal endothelium, and corneal epithelium. The f-actin staining pattern was concentrated at the cell periphery of these four cell types. Vimentin stain was intensely localized in focal areas of the trabecular-meshwork cell, keratocytes, and throughout the corneal cndothelium. A general anticytokeratin antibody was intensely localized in corneal epithelium and endothelium. However, PKK-1 anticytokeratin antibody was seen only in superficial layers of corneal epithelium and not in corneal endothelium. The 4.62 anticytokeratin antibody was not observed in either corneal epithelium or endothelium. None of these three cytokera- tin antibodies were seen in trabccular-mcshwork cells or keratocytes. Desmin stain was not noted in any of these cell types. In general, cytoskeletal staining of unfixed frozen sections showed a similar staining pattern for f-actin and tubulin but a more uniform and intense staining pattern for vimentin and cytokcratin compared with fixed frozen material. The authors conclude that these cytoskclctal stains can differentiate human Irabeciilar-meshwork cells from cells of the cornea in situ. -

Suppression of Keratoepithelin and Myocilin by Small Interfering Rnas (Sirna) in Vitro Ching Yuan University of Minnesota - Twin Cities
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2007 Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro Ching Yuan University of Minnesota - Twin Cities Emily J. Zins University of Minnesota - Twin Cities Abbott .F Clark Alcon Research Ltd. Andrew J.W. Huang Washington University School of Medicine in St. Louis Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Yuan, Ching; Zins, Emily J.; Clark, Abbott .;F and Huang, Andrew J.W., ,"Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro." Molecular Vision.13,. 2083-2095. (2007). https://digitalcommons.wustl.edu/open_access_pubs/1812 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Molecular Vision 2007; 13:2083-95 <http://www.molvis.org/molvis/v13/a236/> ©2007 Molecular Vision Received 25 July 2007 | Accepted 24 October 2007 | Published 7 November 2007 Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro Ching Yuan,1 Emily J. Zins,1 Abbott F. Clark,2 Andrew J.W. Huang1,3 1Department of Ophthalmology, University of Minnesota Minneapolis, MN; 2Alcon Research Ltd., Fort Worth, TX; 3Department of Ophthalmology and Visual Sciences, Washington University, St. Louis, MO Purpose: Mutations of keratoepithelin (KE) and myocilin (MYOC) have been linked to certain types of inherited corneal stromal dystrophy and open-angle glaucoma, respectively. We investigated the potential use of small interfering RNAs (siRNAs) to suppress the expression of KE and MYOC and the related cytotoxicity of mutant myocilins in vitro. -

Decreased Expression of Profilin 2 in Oral Squamous Cell Carcinoma and Its Clinicopathological Implications
ONCOLOGY REPORTS 26: 813-823, 2011 Decreased expression of profilin 2 in oral squamous cell carcinoma and its clinicopathological implications C.Y. MA1,2, C.P. ZHANG1,2, L.P. ZHONG1,2, H.Y. PAN1,2, W.T. CHEN1,2, L.Z. WANG3, O.W. ANDREW4, T. JI1 and W. HAN1,2 1Department of Oral and Maxillofacial Surgery, Ninth People's Hospital, College of Stomatology; 2Shanghai Key Laboratory of Stomatology and Shanghai Research Institute of Stomatology; 3Department of Oral Pathology, Ninth People's Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai 200011, P.R. China; 4Department of Oral and Maxillofacial Surgery, Faculty of Dentistry, National University of Singapore, Singapore 119074, Singapore Received February 8, 2011; Accepted April 11, 2011 DOI: 10.3892/or.2011.1365 Abstract. Profilins are small proteins essential for many clinical and pathological significance. In conclusion, PFN2 normal cellular dynamics and constitute one of the crucial can be utilized as both a potential suppressor marker and a components of actin-based cellular motility. Several recent prognostic protein for OSCC. The function of PFN2 may be to studies have implicated a role for the profilin (PFN) family in regulate the N-WASP/Arp2/3 signaling pathway. cancer pathogenesis and progression. However, their expression and promising functions are largely unknown in oral squamous Introduction cell carcinoma (OSCC). In this study, we analyzed the correlation between PFN1 and PFN2 expression in vitro and Oral squamous cell carcinoma (OSCC) is a significant public in vivo. The protein expression levels were roughly compared health problem with >300,000 new cases being diagnosed between cell lines (HIOEC, HB96) with the employment of annually worldwide (1). -

Genetic Mutations and Mechanisms in Dilated Cardiomyopathy
Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, … , Jessica R. Golbus, Megan J. Puckelwartz J Clin Invest. 2013;123(1):19-26. https://doi.org/10.1172/JCI62862. Review Series Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of congestive heart failure. In hypertrophic cardiomyopathy (HCM), cardiac output is limited by the thickened myocardium through impaired filling and outflow. Mutations in the genes encoding the thick filament components myosin heavy chain and myosin binding protein C (MYH7 and MYBPC3) together explain 75% of inherited HCMs, leading to the observation that HCM is a disease of the sarcomere. Many mutations are “private” or rare variants, often unique to families. In contrast, dilated cardiomyopathy (DCM) is far more genetically heterogeneous, with mutations in genes encoding cytoskeletal, nucleoskeletal, mitochondrial, and calcium-handling proteins. DCM is characterized by enlarged ventricular dimensions and impaired systolic and diastolic function. Private mutations account for most DCMs, with few hotspots or recurring mutations. More than 50 single genes are linked to inherited DCM, including many genes that also link to HCM. Relatively few clinical clues guide the diagnosis of inherited DCM, but emerging evidence supports the use of genetic testing to identify those patients at risk for faster disease progression, congestive heart failure, and arrhythmia. Find the latest version: https://jci.me/62862/pdf Review series Genetic mutations and mechanisms in dilated cardiomyopathy Elizabeth M. McNally, Jessica R. Golbus, and Megan J. Puckelwartz Department of Human Genetics, University of Chicago, Chicago, Illinois, USA. Genetic mutations account for a significant percentage of cardiomyopathies, which are a leading cause of conges- tive heart failure. -

Nei Vision Report.Pdf
VISION RESEARCH A NATIONAL PLAN: 19992003 U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES PUBLIC HEALTH SERVICE NATIONAL INSTITUTES OF HEALTH NATIONAL EYE INSTITUTE A Report of the National Advisory Eye Council DEDICATION Not long after the Roys great intellect, careful experimental approach, creation of the National Eye and keen scientific insights earned him the MERIT Institute (NEI) by Congress Award from the NEI and the Friedenwald Award from in 1968, a young, promising the Association for Research in Vision and vision research scientist Ophthalmology. While maintaining an active and named Roy Steinberg vigorous vision research program, Roy also found time received one of the first to serve as an adviser to the National Institutes of Roy H. Steinberg, M.D., Ph.D. NEI Research Career Health and the NEI. He was a member and later Chair Development Awards. This of the Visual Disorders Study Section, the forerunner marked the beginning of of todays Visual Sciences C. He served as Chair of a long and productive association between the NEI the Retinal Diseases Panel for the NEIs Vision and a researcher who served the vision community ResearchA National Plan: 1987 Evaluation and in many ways. Update and as a consultant to the 19781982 and the 19941998 national plans. He authored the highlights With his great breadth of knowledge and sharp mind, and recommendations from two NEI-sponsored Roy had a clearer grasp than most of the many facets workshopsthe first on the Cell Biology of Retinal of retinal research, both clinical and laboratory. Most Detachment in 1986, and the second on Repair and productive scientists establish a single theme to their Replacement to Restore Sight in 1991. -
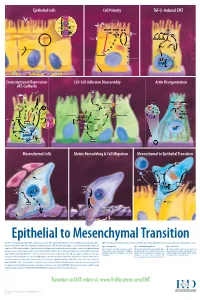
Epithelial to Mesenchymal Transition
Epithelial Cells Cell Polarity TGF-b-Induced EMT MUC-1 O-glycosylation Epithelial Cells ZO-1 Occludin Apical Membrane Tight F-Actin Microvilli Junction Claudin F-Actin p120 β-Catenin Adherens F-Actin Ezrin TGF-β dimer Junction E-Cadherin α-Catenin Plakophilin Crumbs Complex PAR Complex Desmocollin Desmoplakin Desmosome PtdIns(4,5)P2 TGF-β RII TGF-β RI CRB Cdc42Par6 Desmoglein Cytokeratin Pals1 PatJ Tight Junction Plakoglobin aPKC Par3 Domain Smad7 Extracellular PTEN JNK ERK1/2 p38 SARA Smurf1 Cortical Actin Cytoskeleton Space Par3 ZO-1 Adherens Junction PI 3-K Domain Smad-independent Signaling (–) Smad7 Translocation Smad2/3 PtdIns(3,4,5)P3 Smad4 Smad4 NEDD4 Cytokeratin Intermediate Filaments Smad2 Smad4 Smad3 LLGL Proteasome SCRIB DLG Scribble Complex Fibronectin Twist Smad2/3 Vitronectin ZEB 1/2 Microtubule Network Smad4 N-Cadherin Snail Basolateral Membrane CoA, Collagen I Slug CoR MMPs DNA-binding (+) Claudin Desmoplakin Transcription Factor Occludin Cytokeratins E-Cadherin Plakoglobin Integrins β α Nidogen-1/Entactin Perlecan Laminin Collagen IV Transcriptional Repression Cell-Cell Adhesion Disassembly Actin Reorganization of E-Cadherin TGF-β dimer EGF TGF-β RII TGF-β RI IGF FGF Receptor TNF-α Tyrosine Kinase Par6 TNF RI Apical Focal Adhesion Constriction Actin Depolymerization F-Actin Smurf1 Occludin Wnt Frizzled Myosin II Ras RhoA α-Actinin Myosin II ROCK AxinCK1 Dishevelled GSK-3 PI 3-K Src Zyxin MLC Phosphatase APC Proteasome FAK Vinculin RhoA ILK Talin (Inactive) Hakai Talin FAK F-Actin E-Cadherin LIMK Akt Paxillin FAK Stress -

Structural Organization of the Perimysium in Bovine Skeletal Muscle: Junctional Plates and Associated Intracellular Subdomains E
Structural organization of the perimysium in bovine skeletal muscle: Junctional plates and associated intracellular subdomains E. Passerieux, R. Rossignol, A. Chopard, A. Carnino, J.F. Marini, T. Letellier, J.P. Delage To cite this version: E. Passerieux, R. Rossignol, A. Chopard, A. Carnino, J.F. Marini, et al.. Structural organization of the perimysium in bovine skeletal muscle: Junctional plates and associated intracellular subdomains. Journal of Structural Biology, Elsevier, 2006, 154 (2), pp.206 - 216. 10.1016/j.jsb.2006.01.002. hal- 01758589 HAL Id: hal-01758589 https://hal.umontpellier.fr/hal-01758589 Submitted on 4 Apr 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Journal of Structural Biology 154 (2006) 206–216 www.elsevier.com/locate/yjsbi Structural organization of the perimysium in bovine skeletal muscle: Junctional plates and associated intracellular subdomains E. Passerieux a, R. Rossignol a, A. Chopard b, A. Carnino b, J.F. Marini b, a a, T. Letellier , J.P. Delage ¤ a INSERM, U688 Physiopathologie Mitochondriale, Université -

Multiomic Approaches to Uncover the Complexities of Dystrophin-Associated Cardiomyopathy
International Journal of Molecular Sciences Review Multiomic Approaches to Uncover the Complexities of Dystrophin-Associated Cardiomyopathy Aoife Gowran 1,*, Maura Brioschi 2, Davide Rovina 1 , Mattia Chiesa 3,4 , Luca Piacentini 3,* , Sara Mallia 1, Cristina Banfi 2,* , Giulio Pompilio 1,5,6,* and Rosaria Santoro 1,4 1 Unit of Vascular Biology and Regenerative Medicine, Centro Cardiologico Monzino-IRCCS, 20138 Milan, Italy; [email protected] (D.R.); [email protected] (S.M.); [email protected] (R.S.) 2 Unit of Cardiovascular Proteomics, Centro Cardiologico Monzino-IRCCS, 20138 Milan, Italy; [email protected] 3 Bioinformatics and Artificial Intelligence Facility, Centro Cardiologico Monzino-IRCCS, 20138 Milan, Italy; [email protected] 4 Department of Electronics, Information and Biomedical Engineering, Politecnico di Milano, 20133 Milan, Italy 5 Department of Cardiac Surgery, Centro Cardiologico Monzino-IRCCS, 20138 Milan, Italy 6 Department of Biomedical, Surgical and Dental Sciences, University of Milan, 20122 Milan, Italy * Correspondence: [email protected] (A.G.); [email protected] (L.P.); cristina.banfi@cardiologicomonzino.it (C.B.); [email protected] (G.P.) Abstract: Despite major progress in treating skeletal muscle disease associated with dystrophinopathies, cardiomyopathy is emerging as a major cause of death in people carrying dystrophin gene mutations that remain without a targeted cure even with new treatment directions and advances in modelling Citation: Gowran, A.; Brioschi, M.; abilities. The reasons for the stunted progress in ameliorating dystrophin-associated cardiomyopathy Rovina, D.; Chiesa, M.; Piacentini, L.; (DAC) can be explained by the difficulties in detecting pathophysiological mechanisms which can also Mallia, S.; Banfi, C.; Pompilio, G.; Santoro, R. -

Molecular Genetic Analysis of the Adenomatous Polyposis Coli (APC)
Molecular Genetic Analysis of the Adenomatous Polyposis Coli(APC) gene region by Garret Malcolm Hampton A thesis submitted in requirement for the degree of Doctor of Philosophy at the University of London May, 1992 Cancer Genetics Laboratory (formerly Director's Laboratory) Imperial Cancer Research Fund London and Department of Genetics and Biometry University College London London University Page -1- ProQuest Number: 10609182 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a com plete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest ProQuest 10609182 Published by ProQuest LLC(2017). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States C ode Microform Edition © ProQuest LLC. ProQuest LLC. 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106- 1346 To my parents and my wife, Kate Page -2- Abstract Familial Adenomatous Polyposis (FAP) is a rare, autosomal dominant predisposition to colorectal cancer, affecting about one in ten thousand individuals in all populations studied. The gene responsible for this syndrome, designated APC (for Adenomatous Polyposis Coli) was mapped to 5q21-q22 by linkage analysis following a cytogenetic report of a male patient with polyposis and an interstitial deletion on 5q. The high incidence of allele loss at 5q21-q22 in carcinomas of sporadic patients suggests that mutation of the APC gene is a very frequent step in the tumorigenic pathway to nonfamilial colorectal carcinomas and emphasises the importance of isolating the gene and identifying its function.