Rho小g蛋白相关激酶的结构与功能 闫慧娟 勿呢尔 莫日根 范丽菲* (内蒙古大学生命科学学院, 呼和浩特 010021)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Genome-Wide Sirna Screen for Mediators of NF-Κb Activation
Genome-wide siRNA screen for mediators SEE COMMENTARY of NF-κB activation Benjamin E. Gewurza, Fadi Towficb,c,1, Jessica C. Marb,d,1, Nicholas P. Shinnersa,1, Kaoru Takasakia, Bo Zhaoa, Ellen D. Cahir-McFarlanda, John Quackenbushe, Ramnik J. Xavierb,c, and Elliott Kieffa,2 aDepartment of Medicine and Microbiology and Molecular Genetics, Channing Laboratory, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115; bCenter for Computational and Integrative Biology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114; cProgram in Medical and Population Genetics, The Broad Institute of Massachusetts Institute of Technology and Harvard, Cambridge, MA 02142; dDepartment of Biostatistics, Harvard School of Public Health, Boston, MA 02115; and eDepartment of Biostatistics and Computational Biology and Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02115 Contributed by Elliott Kieff, December 16, 2011 (sent for review October 2, 2011) Although canonical NFκB is frequently critical for cell proliferation, (RIPK1). TRADD engages TNFR-associated factor 2 (TRAF2), survival, or differentiation, NFκB hyperactivation can cause malig- which recruits the ubiquitin (Ub) E2 ligase UBC5 and the E3 nant, inflammatory, or autoimmune disorders. Despite intensive ligases cIAP1 and cIAP2. CIAP1/2 polyubiquitinate RIPK1 and study, mammalian NFκB pathway loss-of-function RNAi analyses TRAF2, which recruit and activate the K63-Ub binding proteins have been limited to specific protein classes. We therefore under- TAB1, TAB2, and TAB3, as well as their associated kinase took a human genome-wide siRNA screen for novel NFκB activa- MAP3K7 (TAK1). TAK1 in turn phosphorylates IKKβ activa- tion pathway components. Using an Epstein Barr virus latent tion loop serines to promote IKK activity (4). -

Supplementary Table 1
SI Table S1. Broad protein kinase selectivity for PF-2771. Kinase, PF-2771 % Inhibition at 10 μM Service Kinase, PF-2771 % Inhibition at 1 μM Service rat RPS6KA1 (RSK1) 39 Dundee AURKA (AURA) 24 Invitrogen IKBKB (IKKb) 26 Dundee CDK2 /CyclinA 21 Invitrogen mouse LCK 25 Dundee rabbit MAP2K1 (MEK1) 19 Dundee AKT1 (AKT) 21 Dundee IKBKB (IKKb) 16 Dundee CAMK1 (CaMK1a) 19 Dundee PKN2 (PRK2) 14 Dundee RPS6KA5 (MSK1) 18 Dundee MAPKAPK5 14 Dundee PRKD1 (PKD1) 13 Dundee PIM3 12 Dundee MKNK2 (MNK2) 12 Dundee PRKD1 (PKD1) 12 Dundee MARK3 10 Dundee NTRK1 (TRKA) 12 Invitrogen SRPK1 9 Dundee MAPK12 (p38g) 11 Dundee MAPKAPK5 9 Dundee MAPK8 (JNK1a) 11 Dundee MAPK13 (p38d) 8 Dundee rat PRKAA2 (AMPKa2) 11 Dundee AURKB (AURB) 5 Dundee NEK2 11 Invitrogen CSK 5 Dundee CHEK2 (CHK2) 11 Invitrogen EEF2K (EEF-2 kinase) 4 Dundee MAPK9 (JNK2) 9 Dundee PRKCA (PKCa) 4 Dundee rat RPS6KA1 (RSK1) 8 Dundee rat PRKAA2 (AMPKa2) 4 Dundee DYRK2 7 Dundee rat CSNK1D (CKId) 3 Dundee AKT1 (AKT) 7 Dundee LYN 3 BioPrint PIM2 7 Invitrogen CSNK2A1 (CKIIa) 3 Dundee MAPK15 (ERK7) 6 Dundee CAMKK2 (CAMKKB) 1 Dundee mouse LCK 5 Dundee PIM3 1 Dundee PDPK1 (PDK1) (directed 5 Invitrogen rat DYRK1A (MNB) 1 Dundee RPS6KB1 (p70S6K) 5 Dundee PBK 0 Dundee CSNK2A1 (CKIIa) 4 Dundee PIM1 -1 Dundee CAMKK2 (CAMKKB) 4 Dundee DYRK2 -2 Dundee SRC 4 Invitrogen MAPK12 (p38g) -2 Dundee MYLK2 (MLCK_sk) 3 Invitrogen NEK6 -3 Dundee MKNK2 (MNK2) 2 Dundee RPS6KB1 (p70S6K) -3 Dundee SRPK1 2 Dundee AKT2 -3 Dundee MKNK1 (MNK1) 2 Dundee RPS6KA3 (RSK2) -3 Dundee CHEK1 (CHK1) 2 Invitrogen rabbit MAP2K1 (MEK1) -4 Dundee -

Atu027, a Liposomal Small Interfering RNA Formulation Targeting Protein Kinase N3, Inhibits Cancer Progression
Research Article Atu027, a Liposomal Small Interfering RNA Formulation Targeting Protein Kinase N3, Inhibits Cancer Progression Manuela Aleku,1 Petra Schulz,2 Oliver Keil,1 Ansgar Santel,1 Ute Schaeper,1 Britta Dieckhoff,1 Oliver Janke,1 Jens Endruschat,1 Birgit Durieux,1 Nadine Ro¨der,1 Kathrin Lo¨ffler,1 Christian Lange,1 Melanie Fechtner,1 Kristin Mo¨pert,1 Gerald Fisch,1 Sibylle Dames,1 Wolfgang Arnold,1 Karin Jochims,3 Klaus Giese,1 Bertram Wiedenmann,2 Arne Scholz,2 and Jo¨rg Kaufmann1 1Silence Therapeutics AG; 2Medizinische Klinik mit Schwerpunkt Hepatologie und Gastroenterologie, Charite´-Universita¨tsmedizin, Berlin, Germany; and 3IASON Consulting, Niederzier, Germany Abstract of chemically synthesized siRNA for therapeutic purposes (for a We have previously described a small interfering RNA (siRNA) technology review, see refs. 2, 4, 6–8). With respect to cancer, a delivery system (AtuPLEX) for RNA interference (RNAi) in the variety of nonviral nanoparticles (50–200 nm) have been recently vasculature of mice. Here we report preclinical data for reported to be suitable for RNAi-mediated tumor growth inhibition Atu027, a siRNA-lipoplex directed against protein kinase N3 in experimental mouse models (summarized in ref. 6). In most (PKN3), currently under development for the treatment of cases, various siRNA carriers such as cyclodextrin (transferrin advanced solid cancer. In vitro studies revealed that Atu027- receptor targeted; ref. 9), PEI (RGD-targeted PEG-PEI; ref. 10), mediated inhibition of PKN3 function in primary endothelial DOTAP (self-assembled LPD-nanoparticle; nanosized immunolipo- cells impaired tube formation on extracellular matrix and some; refs. 11, 12), AtuFECT (AtuPLEX; refs. -

Cellular Roles of PKN1
Cellular Roles of PKN1 Neil Torbett This thesis is submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University College University of London August 2003 London Research Institute Cancer Research UK 44 Lincoln’s Inn Fields London W C2A 3PX ProQuest Number: U642486 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. In the unlikely event that the author did not send a complete manuscript and there are missing pages, these will be noted. Also, if material had to be removed, a note will indicate the deletion. uest. ProQuest U642486 Published by ProQuest LLC(2015). Copyright of the Dissertation is held by the Author. All rights reserved. This work is protected against unauthorized copying under Title 17, United States Code. Microform Edition © ProQuest LLC. ProQuest LLC 789 East Eisenhower Parkway P.O. Box 1346 Ann Arbor, Ml 48106-1346 A b s t r a c t Protein Kinase Novel 1 (PKN1), which in part resembles yeast PKCs, has been shown to be under the control of Rho GTPases and 3-Phosphoinositide Dependent Kinase 1. Initial studies tested the hypothesis that Rho-PKNI inputs control PKB phosphorylation. Despite a demonstrable intervention in Rho function, no evidence was obtained that indicated a role for Rho-PKN1 in PKB control. In seeking a cellular role for PKN1, it was found that GPP tagged PKN1 has the ability to translocate in a reversible manner to a vesicular compartment following hyperosmotic stress. PKN1 kinase activity is not necessary for this translocation and in fact the PKN inhibitor HA1077 is also shown to induce PKN1 vesicle accumulation. -

Activation of Diverse Signalling Pathways by Oncogenic PIK3CA Mutations
ARTICLE Received 14 Feb 2014 | Accepted 12 Aug 2014 | Published 23 Sep 2014 DOI: 10.1038/ncomms5961 Activation of diverse signalling pathways by oncogenic PIK3CA mutations Xinyan Wu1, Santosh Renuse2,3, Nandini A. Sahasrabuddhe2,4, Muhammad Saddiq Zahari1, Raghothama Chaerkady1, Min-Sik Kim1, Raja S. Nirujogi2, Morassa Mohseni1, Praveen Kumar2,4, Rajesh Raju2, Jun Zhong1, Jian Yang5, Johnathan Neiswinger6, Jun-Seop Jeong6, Robert Newman6, Maureen A. Powers7, Babu Lal Somani2, Edward Gabrielson8, Saraswati Sukumar9, Vered Stearns9, Jiang Qian10, Heng Zhu6, Bert Vogelstein5, Ben Ho Park9 & Akhilesh Pandey1,8,9 The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signalling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signalling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K-enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signalling events and for discovering novel signalling molecules as readouts of pathway activation or potential therapeutic targets. 1 McKusick-Nathans Institute of Genetic Medicine and Department of Biological Chemistry, Johns Hopkins University School of Medicine, 733 North Broadway, BRB 527, Baltimore, Maryland 21205, USA. -

The Kinesin Spindle Protein Inhibitor Filanesib Enhances the Activity of Pomalidomide and Dexamethasone in Multiple Myeloma
Plasma Cell Disorders SUPPLEMENTARY APPENDIX The kinesin spindle protein inhibitor filanesib enhances the activity of pomalidomide and dexamethasone in multiple myeloma Susana Hernández-García, 1 Laura San-Segundo, 1 Lorena González-Méndez, 1 Luis A. Corchete, 1 Irena Misiewicz- Krzeminska, 1,2 Montserrat Martín-Sánchez, 1 Ana-Alicia López-Iglesias, 1 Esperanza Macarena Algarín, 1 Pedro Mogollón, 1 Andrea Díaz-Tejedor, 1 Teresa Paíno, 1 Brian Tunquist, 3 María-Victoria Mateos, 1 Norma C Gutiérrez, 1 Elena Díaz- Rodriguez, 1 Mercedes Garayoa 1* and Enrique M Ocio 1* 1Centro Investigación del Cáncer-IBMCC (CSIC-USAL) and Hospital Universitario-IBSAL, Salamanca, Spain; 2National Medicines Insti - tute, Warsaw, Poland and 3Array BioPharma, Boulder, Colorado, USA *MG and EMO contributed equally to this work ©2017 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.168666 Received: March 13, 2017. Accepted: August 29, 2017. Pre-published: August 31, 2017. Correspondence: [email protected] MATERIAL AND METHODS Reagents and drugs. Filanesib (F) was provided by Array BioPharma Inc. (Boulder, CO, USA). Thalidomide (T), lenalidomide (L) and pomalidomide (P) were purchased from Selleckchem (Houston, TX, USA), dexamethasone (D) from Sigma-Aldrich (St Louis, MO, USA) and bortezomib from LC Laboratories (Woburn, MA, USA). Generic chemicals were acquired from Sigma Chemical Co., Roche Biochemicals (Mannheim, Germany), Merck & Co., Inc. (Darmstadt, Germany). MM cell lines, patient samples and cultures. Origin, authentication and in vitro growth conditions of human MM cell lines have already been characterized (17, 18). The study of drug activity in the presence of IL-6, IGF-1 or in co-culture with primary bone marrow mesenchymal stromal cells (BMSCs) or the human mesenchymal stromal cell line (hMSC–TERT) was performed as described previously (19, 20). -

Inhibition of ERK 1/2 Kinases Prevents Tendon Matrix Breakdown Ulrich Blache1,2,3, Stefania L
www.nature.com/scientificreports OPEN Inhibition of ERK 1/2 kinases prevents tendon matrix breakdown Ulrich Blache1,2,3, Stefania L. Wunderli1,2,3, Amro A. Hussien1,2, Tino Stauber1,2, Gabriel Flückiger1,2, Maja Bollhalder1,2, Barbara Niederöst1,2, Sandro F. Fucentese1 & Jess G. Snedeker1,2* Tendon extracellular matrix (ECM) mechanical unloading results in tissue degradation and breakdown, with niche-dependent cellular stress directing proteolytic degradation of tendon. Here, we show that the extracellular-signal regulated kinase (ERK) pathway is central in tendon degradation of load-deprived tissue explants. We show that ERK 1/2 are highly phosphorylated in mechanically unloaded tendon fascicles in a vascular niche-dependent manner. Pharmacological inhibition of ERK 1/2 abolishes the induction of ECM catabolic gene expression (MMPs) and fully prevents loss of mechanical properties. Moreover, ERK 1/2 inhibition in unloaded tendon fascicles suppresses features of pathological tissue remodeling such as collagen type 3 matrix switch and the induction of the pro-fbrotic cytokine interleukin 11. This work demonstrates ERK signaling as a central checkpoint to trigger tendon matrix degradation and remodeling using load-deprived tissue explants. Tendon is a musculoskeletal tissue that transmits muscle force to bone. To accomplish its biomechanical function, tendon tissues adopt a specialized extracellular matrix (ECM) structure1. Te load-bearing tendon compart- ment consists of highly aligned collagen-rich fascicles that are interspersed with tendon stromal cells. Tendon is a mechanosensitive tissue whereby physiological mechanical loading is vital for maintaining tendon archi- tecture and homeostasis2. Mechanical unloading of the tissue, for instance following tendon rupture or more localized micro trauma, leads to proteolytic breakdown of the tissue with severe deterioration of both structural and mechanical properties3–5. -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain -
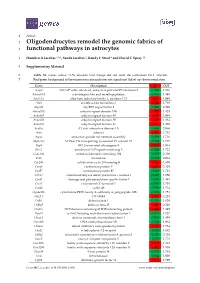
Oligodendrocytes Remodel the Genomic Fabrics of Functional Pathways in Astrocytes
1 Article 2 Oligodendrocytes remodel the genomic fabrics of 3 functional pathways in astrocytes 4 Dumitru A Iacobas 1,2,*, Sanda Iacobas 3, Randy F Stout 4 and David C Spray 2,5 5 Supplementary Material 6 Table S1. Genes whose >1.5x absolute fold-change did not meet the individual CUT criterion. 7 Red/green background of the expression ratio indicates not significant (false) up-/down-regulation. Gene Description X CUT Acap2 ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 -1.540 1.816 Adamts18 a disintegrin-like and metallopeptidase -1.514 1.594 Akr1c12 aldo-keto reductase family 1, member C12 1.866 1.994 Alx3 aristaless-like homeobox 3 1.536 1.769 Alyref2 Aly/REF export factor 2 -1.880 2.208 Ankrd33b ankyrin repeat domain 33B 1.593 1.829 Ankrd45 ankyrin repeat domain 45 1.514 1.984 Ankrd50 ankyrin repeat domain 50 1.628 1.832 Ankrd61 ankyrin repeat domain 61 1.645 1.802 Arid1a AT rich interactive domain 1A -1.668 2.066 Artn artemin 1.524 1.732 Aspm abnormal spindle microtubule assembly -1.693 1.716 Atp6v1e1 ATPase, H+ transporting, lysosomal V1 subunit E1 -1.679 1.777 Bag4 BCL2-associated athanogene 4 1.723 1.914 Birc3 baculoviral IAP repeat-containing 3 -1.588 1.722 Ccdc104 coiled-coil domain containing 104 -1.819 2.130 Ccl2 chemokine -1.699 2.034 Cdc20b cell division cycle 20 homolog B 1.512 1.605 Cenpf centromere protein F 2.041 2.128 Cep97 centrosomal protein 97 -1.641 1.723 COX1 mitochondrially encoded cytochrome c oxidase I -1.607 1.650 Cpsf7 cleavage and polyadenylation specific factor 7 -1.635 1.891 Crct1 cysteine-rich -

Protein Kinase N1 Promotes Proliferation and Invasion of Liver Cancer
EXPERIMENTAL AND THERAPEUTIC MEDICINE 21: 651, 2021 Protein kinase N1 promotes proliferation and invasion of liver cancer XIA WANG1, YANSONG GE2, MINGQI SHI2, HANHAN DAI2, WEI LIU2 and PEIYUAN WANG2 1Department of Pathology, Binzhou Medical University, Yantai, Shandong 264003; 2Department of Radiology, Yantai Affiliated Hospital of Binzhou Medical University, Yantai, Shandong 264100, P.R. China Received March 11, 2020; Accepted March 22, 2021 DOI: 10.3892/etm.2021.10083 Abstract. Protein kinase (PK) N1, also called PKC‑related accounts for ~55%, with 110,000 deaths every year (1‑3). protein 1, participates in the proliferation, invasion and Currently, there are a variety of treatment options for liver metastasis of various malignant tumors. However, the role of cancer at different stages, including routine options (such PKN1 in liver cancer remains to be elucidated. The present as hepatectomy and transcatheter arterial chemoemboliza‑ study investigated the expression of PKN1 using immuno‑ tion), molecular targeted therapy (VEGF/VEGF monoclonal histochemistry in surgical specimens from 36 patients and antibody, EGF receptor inhibitors) and immunotherapy analyzed the correlation with VEGF, microvascular density (programmed death‑1/programmed death‑ligand 1), which can (MVD), cell proliferation index (Ki67) and clinicopatholog‑ prolong the survival time and prognosis of some patients (4‑6). ical parameters. PKN1 was highly expressed in hepatocellular However, due to the selection of indications, sensitivity to carcinoma (HCC) and was positively correlated with histo‑ treatment and other reasons, most patients have not received logical grading of HCC, Ki67 expression and MVD. PKN1 effective treatment. Therefore, it is important to explore the expression in moderately and poorly differentiated HCC was mechanism of proliferation, invasion and metastasis of liver significantly higher compared with highly differentiated HCC. -

Hypoxia Transcriptomic Modifications Induced by Proton Irradiation In
Journal of Personalized Medicine Article Hypoxia Transcriptomic Modifications Induced by Proton Irradiation in U87 Glioblastoma Multiforme Cell Line Valentina Bravatà 1,2,† , Walter Tinganelli 3,†, Francesco P. Cammarata 1,2,* , Luigi Minafra 1,2,* , Marco Calvaruso 1,2 , Olga Sokol 3 , Giada Petringa 2, Giuseppe A.P. Cirrone 2, Emanuele Scifoni 4 , Giusi I. Forte 1,2,‡ and Giorgio Russo 1,2,‡ 1 Institute of Molecular Bioimaging and Physiology–National Research Council (IBFM-CNR), 90015 Cefalù, Italy; [email protected] (V.B.); [email protected] (M.C.); [email protected] (G.I.F.); [email protected] (G.R.) 2 Laboratori Nazionali del SUD, Istituto Nazionale di Fisica Nucleare (LNS-INFN), 95100 Catania, Italy; [email protected] (G.P.); [email protected] (G.A.P.C.) 3 Biophysics Department, GSI Helmholtzzentrum für Schwerionenforschung GmbH, 64291 Darmstadt, Germany; [email protected] (W.T.); [email protected] (O.S.) 4 Trento Institute for Fundamental Physics and Applications (TIFPA), Istituto Nazionale Fisica Nucleare (INFN), 38123 Trento, Italy; [email protected] * Correspondence: [email protected] (F.P.C.); [email protected] (L.M.) † These authors contributed equally to this work. ‡ These authors contributed equally to this work. Abstract: In Glioblastoma Multiforme (GBM), hypoxia is associated with radioresistance and poor prognosis. Since standard GBM treatments are not always effective, new strategies are needed Citation: Bravatà, V.; Tinganelli, W.; to overcome resistance to therapeutic treatments, including radiotherapy (RT). Our study aims Cammarata, F.P.; Minafra, L.; to shed light on the biomarker network involved in a hypoxic (0.2% oxygen) GBM cell line that Calvaruso, M.; Sokol, O.; Petringa, G.; is radioresistant after proton therapy (PT).