Validation List No. 85
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Geomicrobiological Processes in Extreme Environments: a Review
202 Articles by Hailiang Dong1, 2 and Bingsong Yu1,3 Geomicrobiological processes in extreme environments: A review 1 Geomicrobiology Laboratory, China University of Geosciences, Beijing, 100083, China. 2 Department of Geology, Miami University, Oxford, OH, 45056, USA. Email: [email protected] 3 School of Earth Sciences, China University of Geosciences, Beijing, 100083, China. The last decade has seen an extraordinary growth of and Mancinelli, 2001). These unique conditions have selected Geomicrobiology. Microorganisms have been studied in unique microorganisms and novel metabolic functions. Readers are directed to recent review papers (Kieft and Phelps, 1997; Pedersen, numerous extreme environments on Earth, ranging from 1997; Krumholz, 2000; Pedersen, 2000; Rothschild and crystalline rocks from the deep subsurface, ancient Mancinelli, 2001; Amend and Teske, 2005; Fredrickson and Balk- sedimentary rocks and hypersaline lakes, to dry deserts will, 2006). A recent study suggests the importance of pressure in the origination of life and biomolecules (Sharma et al., 2002). In and deep-ocean hydrothermal vent systems. In light of this short review and in light of some most recent developments, this recent progress, we review several currently active we focus on two specific aspects: novel metabolic functions and research frontiers: deep continental subsurface micro- energy sources. biology, microbial ecology in saline lakes, microbial Some metabolic functions of continental subsurface formation of dolomite, geomicrobiology in dry deserts, microorganisms fossil DNA and its use in recovery of paleoenviron- Because of the unique geochemical, hydrological, and geological mental conditions, and geomicrobiology of oceans. conditions of the deep subsurface, microorganisms from these envi- Throughout this article we emphasize geomicrobiological ronments are different from surface organisms in their metabolic processes in these extreme environments. -

Insights Into Archaeal Evolution and Symbiosis from the Genomes of a Nanoarchaeon and Its Inferred Crenarchaeal Host from Obsidian Pool, Yellowstone National Park
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Microbiology Publications and Other Works Microbiology 4-22-2013 Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar University of Tennessee - Knoxville, [email protected] Kira S. Makarova National Institutes of Health David E. Graham University of Tennessee - Knoxville, [email protected] Yuri I. Wolf National Institutes of Health Eugene V. Koonin National Institutes of Health See next page for additional authors Follow this and additional works at: https://trace.tennessee.edu/utk_micrpubs Part of the Microbiology Commons Recommended Citation Biology Direct 2013, 8:9 doi:10.1186/1745-6150-8-9 This Article is brought to you for free and open access by the Microbiology at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Microbiology Publications and Other Works by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. Authors Mircea Podar, Kira S. Makarova, David E. Graham, Yuri I. Wolf, Eugene V. Koonin, and Anna-Louise Reysenbach This article is available at TRACE: Tennessee Research and Creative Exchange: https://trace.tennessee.edu/ utk_micrpubs/44 Podar et al. Biology Direct 2013, 8:9 http://www.biology-direct.com/content/8/1/9 RESEARCH Open Access Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park Mircea Podar1,2*, Kira S Makarova3, David E Graham1,2, Yuri I Wolf3, Eugene V Koonin3 and Anna-Louise Reysenbach4 Abstract Background: A single cultured marine organism, Nanoarchaeum equitans, represents the Nanoarchaeota branch of symbiotic Archaea, with a highly reduced genome and unusual features such as multiple split genes. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Brian W. Waters
INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS (Under the direction of Dr. William B. Whitman) ABSTRACT With the emergence of Methanococcus maripaludis as a genetic model for methanogens, it becomes imperative to devise inexpensive yet effective ways to grow large numbers of cells for protein studies. Chapter 2 details methods that were used to cut costs of growing M. maripaludis in large scale. Also, large scale growth under different conditions was explored in order to find the conditions that yield the most cells. Chapter 3 details the phylogenetic analysis of 2-oxoacid oxidoreductase (OR) homologs from M. maripaludis. ORs are enzymes that catalyze the oxidative decarboxylation of 2-oxoacids to their acyl-CoA derivatives in many prokaryotes. Also in chapter 3, a specific OR homolog in M. maripaludis, an indolepyruvate oxidoreductase (IOR), was mutagenized, and the mutant was characterized. PCR and Southern hybridization analysis showed gene replacement. A no-growth phenotype on media containing aromatic amino acid derivatives was found. INDEX WORDS: Methanococcus maripaludis, fermentor, 2-oxoacid oxidoreductase, indolepyruvate oxidoreductase INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. WATERS B.S., The University of Georgia,1999 A Thesis Submitted to the Graduate Faculty of The University of Georgia in Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE ATHENS, GEORGIA 2002 © 2002 Brian W. Waters All Rights Reserved INVESTIGATION OF 2-OXOACID OXIDOREDUCTASES IN METHANOCOCCUS MARIPALUDIS AND LARGE-SCALE GROWTH OF M. MARIPALUDIS by BRIAN W. -

26Sor Em2rray /Mmucd
D4006- Gut Microbiota Analysis UC Davis MMPC - Microbiome & Host Response Core Contents 1 Methods: 1 1.1 Sequencing . .1 1.2 Data processing . .1 2 Summary of Findings: 2 2.1 Sequencing analysis . .2 2.2 Microbial diversity . .2 2.2.1 Alpha Diversity . .2 2.2.2 Beta Diversity . 10 2.3 Data analysis using taxa abundance data . 13 2.3.1 Stacked bar graphs of Taxa abundances at each level . 14 A Appendix 1 (Taxa Abundance Tables) 30 B Appendix 2 (Taxa removed from filtered datasets at each level) 37 References: 43 Core Contacts: Helen E. Raybould, Ph.D., Core Leader ([email protected]) Trina A. Knotts, Ph.D., Core Co-Leader ([email protected]) Michael L. Goodson, Ph.D., Core Scientist ([email protected]) Client(s): Kent Lloyd, DVM PhD ;MMRRC; UC Davis Project #: MBP-2079 MMRRC strain ID: MMRRC_043514 Animal Information: The strain was donated to the MMRRC by Russell Ray at Baylor College of Medicine. Fecal samples were obtained from animals housed under the care of Russell Ray at Baylor College of Medicine. 1 Methods: Brief Project Description: MMRRC strains are often contributed to the MMRRC to fulfill the resource sharing aspects of NIH grants. Since transporting mice to another facilty often causes a microbiota shift, having a record of the original fecal microbiota from the donor institution where the original phenotyping or testing was performed may prove helpful if a phenotype is lost after transfer. Several MMRRC mouse lines were selected for fecal microbiota profiling of the microbiota. Table 1: Animal-Strain Information X.SampleID TreatmentGroup Animal_ID Genotype Line Sex MMRRC.043514.Mx054 MMRRC.043514_Het_M Mx054 Het MMRRC.043514 M MMRRC.043514.Mx415 MMRRC.043514_Het_M Mx415 Het MMRRC.043514 M 1 1.1 Sequencing Frozen fecal or regional gut samples were shipped on dry ice to UC Davis MMPC and Host Microbe Systems Biology Core. -
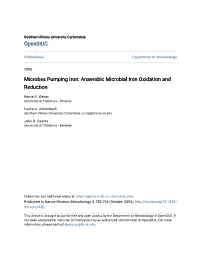
Fe(II) Fe(III)
Southern Illinois University Carbondale OpenSIUC Publications Department of Microbiology 2006 Microbes Pumping Iron: Anaerobic Microbial Iron Oxidation and Reduction Karrie A. Weber University of California - Berkeley Laurie A. Achenbach Southern Illinois University Carbondale, [email protected] John D. Coates University of California - Berkeley Follow this and additional works at: https://opensiuc.lib.siu.edu/micro_pubs Published in Nature Reviews Microbiology 4, 752-764 (October 2006), http://dx.doi.org/10.1038/ nrmicro1490. This Article is brought to you for free and open access by the Department of Microbiology at OpenSIUC. It has been accepted for inclusion in Publications by an authorized administrator of OpenSIUC. For more information, please contact [email protected]. 1 Microbes Pumping Iron: 2 Anaerobic Microbial Iron Oxidation and Reduction 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Karrie A. Weber1, Laurie A. Achenbach2, and John D. Coates1* 18 19 20 21 22 23 1Department of Plant and Microbial Biology 24 271 Koshland Hall 25 University of California, Berkeley 26 Berkeley, CA 94720 27 28 29 2Department of Microbiology 30 Southern Illinois University 31 Carbondale, IL 62901 32 33 34 35 Corresponding Author 36 Tel: (510) 643-8455 37 Fax: (510) 642-4995 38 e-mail: [email protected] 39 1 1 Abstract 2 Iron (Fe) has long been a recognized physiological requirement for life, yet its role for a 3 broad diversity of microorganisms persisting in water, soils, and sediments extends well-beyond 4 a nutritional necessity. Microorganisms from within both the domain Archaea and Bacteria are 5 capable of metabolically exploiting the favorable redox potential between the Fe(III)/Fe(II) 6 couple. -

Supplementary Material Impacts of Radiation on the Bacterial And
Supplementary Material Impacts of radiation on the bacterial and fungal microbiome of small mammals in the Chernobyl Exclusion Zone Rachael E. Antwis, Nicholas A. Beresford, Joseph A. Jackson, Ross Fawkes, Catherine L. Barnett, Elaine Potter, Lee Walker, Sergey Gaschak, Michael D. Wood Table S1 Sample sizes of host species used in the study, and associated sex, total absorbed dose rate, burn and site categories for these samples. Animals estimated to receive total absorbed dose rates of <4 µGy h-1 were assigned ‘low’, those with estimated dose rates of 4-42 µGy h-1 assigned ‘medium’, and those >42 µGy h-1 assigned to the ‘high’ category. All samples from 2017 were collected within the Red Forest but from areas that had experienced different degrees of damage from forest fires. Samples from 2018 were either collected within or outside the Red Forest. Sampling information Sex Total absorbed dose category Burn category Site category Year Sample Host species Total n Female Male Unidentified Low Medium High Burnt Burnt Unburnt Inside Red Outside type (regrowth) (minimal Forest Red regrowth) Forest 2017 Faeces Bank vole 22 5 16 1 0 0 22 3 1 18 All - 2017 Faeces Striped field mouse 29 10 18 1 0 7 22 15 0 14 All - 2017 Faeces Wood mouse 27 6 20 1 0 0 27 2 25 0 All - 2017 Faeces Yellow-necked mouse 58 28 29 1 0 7 51 19 17 22 All - 2018 Gut Bank vole 142 59 83 0 42 64 36 - - - 34 108 (caecum) RESULTS Differences in community composition between host species and sample types Wood mice and yellow-necked mice had a similar bacterial community composition to one another (Figure 1a), with the exception of the family Helicobacteraceae (Z = 2.982, p = 0.023; Figure 1c). -

The Histology, Microbiology, and Molecular Ecology of Tissue-Loss Diseases Affecting Acropora Cervicornis in the Upper Florida Keys
THE HISTOLOGY, MICROBIOLOGY, AND MOLECULAR ECOLOGY OF TISSUE-LOSS DISEASES AFFECTING ACROPORA CERVICORNIS IN THE UPPER FLORIDA KEYS by Katheryn W. Patterson A Dissertation Submitted to the Graduate Faculty of George Mason University in Partial Fulfillment of The Requirements for the Degree of Doctor of Philosophy Environmental Science and Public Policy Committee: _________________________________________ Dr. Esther C. Peters, Dissertation Director _________________________________________ Dr. Cara Frankenfeld, Committee Member _________________________________________ Dr. Patrick Gillevet, Committee Member _________________________________________ Dr. Robert Jonas, Committee Member _________________________________________ Dr. E.C.M. Parsons, Committee Member _________________________________________ Dr. Albert Torzilli, Graduate Program Director _________________________________________ Dr. Robert Jonas, Department Chairperson _________________________________________ Dr. Donna M. Fox, Associate Dean, Office of Student Affairs & Special Programs, College of Science _________________________________________ Dr. Peggy Agouris, Dean, College of Science Date: __________________________________ Summer Semester 2015 George Mason University Fairfax, VA The Histology, MicrobiOlogy, and Molecular Ecology of Tissue-Loss Diseases Affecting Acropora cervicornis in the Upper Florida Keys A Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy at George Mason University by Katheryn W. Patterson Master of Science George Mason University, 2010 Director: Esther C. Peters, Professor Department of Environmental Science and Public Policy Summer Semester 2015 George Mason University Fairfax, VA This work is licensed under a creative commons attribution-noderivs 3.0 unported license. ii DEDICATION This dissertation is in memory of and dedicated to two professors, mentors, and friends that helped inspire and challenge me to be the person I am today. Thank you for encouraging me to stay on the path to become a marine biologist, Dr. -

Fmicb-08-02642
Edinburgh Research Explorer Identification, Comparison, and Validation of Robust Rumen Microbial Biomarkers for Methane Emissions Using DiverseBreeds and Basal Diets Citation for published version: Auffret, MD, Stewart, R, Dewhurst, R, Duthie, C-A, Rooke, JA, Wallace, R, Freeman, T, Snelling, TJ, Watson, M & Roehe, R 2018, 'Identification, Comparison, and Validation of Robust Rumen Microbial Biomarkers for Methane Emissions Using DiverseBreeds and Basal Diets', Frontiers in Microbiology, vol. 8, 2642. https://doi.org/10.3389/fmicb.2017.02642, https://doi.org/10.3389/fmicb.2017.02642 Digital Object Identifier (DOI): 10.3389/fmicb.2017.02642 10.3389/fmicb.2017.02642 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Frontiers in Microbiology Publisher Rights Statement: This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms. General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. -

Bibliography
Bibliography Abella, C.A., X.P. Cristina, A. Martinez, I. Pibernat and X. Vila. 1998. on moderate concentrations of acetate: production of single cells. Two new motile phototrophic consortia: "Chlorochromatium lunatum" Appl. Microbiol. Biotechnol. 35: 686-689. and "Pelochromatium selenoides". Arch. Microbiol. 169: 452-459. Ahring, B.K, P. Westermann and RA. Mah. 1991b. Hydrogen inhibition Abella, C.A and LJ. Garcia-Gil. 1992. Microbial ecology of planktonic of acetate metabolism and kinetics of hydrogen consumption by Me filamentous phototrophic bacteria in holomictic freshwater lakes. Hy thanosarcina thermophila TM-I. Arch. Microbiol. 157: 38-42. drobiologia 243-244: 79-86. Ainsworth, G.C. and P.H.A Sheath. 1962. Microbial Classification: Ap Acca, M., M. Bocchetta, E. Ceccarelli, R Creti, KO. Stetter and P. Cam pendix I. Symp. Soc. Gen. Microbiol. 12: 456-463. marano. 1994. Updating mass and composition of archaeal and bac Alam, M. and D. Oesterhelt. 1984. Morphology, function and isolation terial ribosomes. Archaeal-like features of ribosomes from the deep of halobacterial flagella. ]. Mol. Biol. 176: 459-476. branching bacterium Aquifex pyrophilus. Syst. Appl. Microbiol. 16: 629- Albertano, P. and L. Kovacik. 1994. Is the genus LeptolynglYya (Cyano 637. phyte) a homogeneous taxon? Arch. Hydrobiol. Suppl. 105: 37-51. Achenbach-Richter, L., R Gupta, KO. Stetter and C.R Woese. 1987. Were Aldrich, H.C., D.B. Beimborn and P. Schönheit. 1987. Creation of arti the original eubacteria thermophiles? Syst. Appl. Microbiol. 9: 34- factual internal membranes during fixation of Methanobacterium ther 39. moautotrophicum. Can.]. Microbiol. 33: 844-849. Adams, D.G., D. Ashworth and B. -
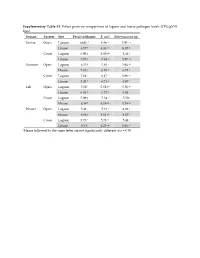
Supplementary Table S1. Fisher Pairwise Comparisons of Lagoon and House Pathogen Levels (CFU/Gvss Log10) Season System Site Fecal Coliforms E
Supplementary Table S1. Fisher pairwise comparisons of lagoon and house pathogen levels (CFU/gVSS log10) Season System Site Fecal coliforms E. coli Enterococcus sp. Spring Open Lagoon 6.00 g1 5.46 hi 5.91 cd House 6.57 e 6.33 cd 6.35 b Cover Lagoon 6.00 g 5.60 gh 5.44 f House 7.73 a 7.63 a 5.87 cd Summer Open Lagoon 6.35 f 5.83 f 5.94 cd House 7.83 a 6.10 e 6.73 a Cover Lagoon 7.04 c 6.47 c 5.86 cd House 7.42 b 6.71 b 6.07 c Fall Open Lagoon 5.58 i 5.58 gh 5.56 ef House 6.81 d 6.72 b 6.01 c Cover Lagoon 5.99 g 5.34 i 5.50 f House 6.38 f 6.19 de 5.74 de Winter Open Lagoon 5.41 j 5.13 j 4.88 g House 6.06 g 5.61 gh 6.67 a Cover Lagoon 5.73 h 5.73 fg 5.44 f House 6.34 f 6.25 de 5.86 cd 1Means followed by the same letter are not significantly different at p = 0.05 Supplementary Table S2. Relative abundances of OTUs identified using universal bacterial primer set, presented as relative abundances (%) Only bacterial families are counted in Figure 5 and discussion involving bacterial family identification. # Identified in # Identified from all 8 All 16 All #OTU Table SpOLRA SuOLRA FOLRA WOLRA SpCLRA SuCLRA FCLRA WCLRA SpOHRA SuOHRA FOHRA WOHRA SpCHRA SuCHRA FCHRA WCHRA all samples lagoon samples Samples Lagoons Notes Key Patulibacteraceae 0 9.93739E‐05* 0.000134 0.000116 0 0 0 0 0 0 0.000452 0 0 0 0 0 4 3 Sp = Spring O = Open Ruaniaceae 0 0 0 0 0 0 0 0 0 0 0 0 0 0.000221577 0 7.73E‐05 2 0 Su = Summer C = Covered Thermoactinomycetaceae 0 0 0.000267 0.000116 0 0.00037092 0.000529 0.000526 0 0.000342922 0.000271 0.000197 0 0 0 0 8 5 F = Fall L = Lagoon Beutenbergiaceae 0.000226334 0.000331246 0.000579 0.001663 0.000620176 0 0.000106 0.000807 0.002320743 0.000571537 0.000723 0.000591 0.000169731 0 0.000606 0 13 7 W = Winter H = House Thermoanaerobacterales Family III. -

Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes
GBE Different Ways of Doing the Same: Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes Dennifier Costa Brandao~ Cruz1, Lenon Lima Santana1, Alexandre Siqueira Guedes2, Jorge Teodoro de Souza3,*, and Phellippe Arthur Santos Marbach1,* 1CCAAB, Biological Sciences, Recoˆ ncavo da Bahia Federal University, Cruz das Almas, Bahia, Brazil 2Agronomy School, Federal University of Goias, Goiania,^ Goias, Brazil 3 Department of Phytopathology, Federal University of Lavras, Minas Gerais, Brazil Downloaded from https://academic.oup.com/gbe/article/11/4/1235/5345563 by guest on 27 September 2021 *Corresponding authors: E-mails: [email protected]fla.br; [email protected]. Accepted: February 16, 2019 Abstract The last two steps of the purine biosynthetic pathway may be catalyzed by different enzymes in prokaryotes. The genes that encode these enzymes include homologs of purH, purP, purO and those encoding the AICARFT and IMPCH domains of PurH, here named purV and purJ, respectively. In Bacteria, these reactions are mainly catalyzed by the domains AICARFT and IMPCH of PurH. In Archaea, these reactions may be carried out by PurH and also by PurP and PurO, both considered signatures of this domain and analogous to the AICARFT and IMPCH domains of PurH, respectively. These genes were searched for in 1,403 completely sequenced prokaryotic genomes publicly available. Our analyses revealed taxonomic patterns for the distribution of these genes and anticorrelations in their occurrence. The analyses of bacterial genomes revealed the existence of genes coding for PurV, PurJ, and PurO, which may no longer be considered signatures of the domain Archaea. Although highly divergent, the PurOs of Archaea and Bacteria show a high level of conservation in the amino acids of the active sites of the protein, allowing us to infer that these enzymes are analogs.