Metabolic Reactions Containing Cdegs in the Genome-Based Mouse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Aldehyde Dehydrogenase ALDH2*2 Allele Exhibits Dominance Over ALDH2*1 in Transduced Hela Cells
The aldehyde dehydrogenase ALDH2*2 allele exhibits dominance over ALDH2*1 in transduced HeLa cells. Q Xiao, … , T Johnston, D W Crabb J Clin Invest. 1995;96(5):2180-2186. https://doi.org/10.1172/JCI118272. Research Article Individuals heterozygous or homozygous for the variant aldehyde dehydrogenase (ALDH2) allele (ALDH2*2), which encodes a protein differing only at residue 487 from the normal protein, have decreased ALDH2 activity in liver extracts and experience cutaneous flushing when they drink alcohol. The mechanisms by which this allele exerts its dominant effect is unknown. To study this effect, the human ALDH2*1 cDNA was cloned and the ALDH2*2 allele was generated by site-directed mutagenesis. These cDNAs were transduced using retroviral vectors into HeLa and CV1 cells, which do not express ALDH2. The normal allele directed synthesis of immunoreactive ALDH2 protein (ALDH2E) with the expected isoelectric point. Extracts of these cells contained increased aldehyde dehydrogenase activity with low Km for the aldehyde substrate. The ALDH2*2 allele directed synthesis of mRNA and immunoreactive protein (ALDH2K), but the protein lacked enzymatic activity. When ALDH2*1-expressing cells were transduced with ALDH2*2 vectors, both mRNAs were expressed and immunoreactive proteins with isoelectric points ranging between those of ALDH2E and ALDH2K were present, indicating that the subunits formed heteromers. ALDH2 activity in these cells was reduced below that of the parental ALDH2*1-expressing cells. Thus, the ALDH2*2 allele is sufficient to cause ALDH2 deficiency in vitro. Find the latest version: https://jci.me/118272/pdf The Aldehyde Dehydrogenase ALDH2*2 Allele Exhibits Dominance over ALDH2*1 in Transduced HeLa Cells Qing Xiao, * Henry Weiner,* Timothy Johnston,* and David W. -

Phosphodiesterase 1B Knock-Out Mice Exhibit Exaggerated Locomotor Hyperactivity and DARPP-32 Phosphorylation in Response to Dopa
The Journal of Neuroscience, June 15, 2002, 22(12):5188–5197 Phosphodiesterase 1B Knock-Out Mice Exhibit Exaggerated Locomotor Hyperactivity and DARPP-32 Phosphorylation in Response to Dopamine Agonists and Display Impaired Spatial Learning Tracy M. Reed,1,3 David R. Repaske,2* Gretchen L. Snyder,4 Paul Greengard,4 and Charles V. Vorhees1* Divisions of 1Developmental Biology and 2Endocrinology, Children’s Hospital Research Foundation, Cincinnati, Ohio 45229, 3Department of Biology, College of Mount St. Joseph, Cincinnati, Ohio 45233, and 4Laboratory of Molecular and Cellular Neuroscience, Rockefeller University, New York, New York 10021 Using homologous recombination, we generated mice lack- maze spatial-learning deficits. These results indicate that en- ing phosphodiesterase-mediated (PDE1B) cyclic nucleotide- hancement of cyclic nucleotide signaling by inactivation of hydrolyzing activity. PDE1B Ϫ/Ϫ mice showed exaggerated PDE1B-mediated cyclic nucleotide hydrolysis plays a signifi- hyperactivity after acute D-methamphetamine administra- cant role in dopaminergic function through the DARPP-32 and tion. Striatal slices from PDE1B Ϫ/Ϫ mice exhibited increased related transduction pathways. levels of phospho-Thr 34 DARPP-32 and phospho-Ser 845 Key words: phosphodiesterases; DARPP-32; dopamine- GluR1 after dopamine D1 receptor agonist or forskolin stimu- stimulated locomotor activity; spatial learning and memory; lation. PDE1B Ϫ/Ϫ and PDE1B ϩ/Ϫ mice demonstrated Morris Morris water maze; methamphetamine; SKF81297; forskolin Calcium/calmodulin-dependent phosphodiesterases (CaM- (CaMKII) and calcineurin and have the potential to activate PDEs) are members of one of 11 families of PDEs (Soderling et CaM-PDEs. Dopamine D1 or D2 receptor activation leads to al., 1999;Yuasa et al., 2001) and comprise the only family that acts adenylyl cyclase activation or inhibition, respectively (Traficante ϩ as a potential point of interaction between the Ca 2 and cyclic et al., 1976; Monsma et al., 1990; Cunningham and Kelley, 1993; nucleotide signaling pathways. -

N-Glycan Trimming in the ER and Calnexin/Calreticulin Cycle
Neurotransmitter receptorsGABA and A postsynapticreceptor activation signal transmission Ligand-gated ion channel transport GABAGABA Areceptor receptor alpha-5 alpha-1/beta-1/gamma-2 subunit GABA A receptor alpha-2/beta-2/gamma-2GABA receptor alpha-4 subunit GABAGABA receptor A receptor beta-3 subunitalpha-6/beta-2/gamma-2 GABA-AGABA receptor; A receptor alpha-1/beta-2/gamma-2GABA receptoralpha-3/beta-2/gamma-2 alpha-3 subunit GABA-A GABAreceptor; receptor benzodiazepine alpha-6 subunit site GABA-AGABA-A receptor; receptor; GABA-A anion site channel (alpha1/beta2 interface) GABA-A receptor;GABA alpha-6/beta-3/gamma-2 receptor beta-2 subunit GABAGABA receptorGABA-A receptor alpha-2receptor; alpha-1 subunit agonist subunit GABA site Serotonin 3a (5-HT3a) receptor GABA receptorGABA-C rho-1 subunitreceptor GlycineSerotonin receptor subunit3 (5-HT3) alpha-1 receptor GABA receptor rho-2 subunit GlycineGlycine receptor receptor subunit subunit alpha-2 alpha-3 Ca2+ activated K+ channels Metabolism of ingested SeMet, Sec, MeSec into H2Se SmallIntermediateSmall conductance conductance conductance calcium-activated calcium-activated calcium-activated potassium potassium potassiumchannel channel protein channel protein 2 protein 1 4 Small conductance calcium-activatedCalcium-activated potassium potassium channel alpha/beta channel 1 protein 3 Calcium-activated potassiumHistamine channel subunit alpha-1 N-methyltransferase Neuraminidase Pyrimidine biosynthesis Nicotinamide N-methyltransferase Adenosylhomocysteinase PolymerasePolymeraseHistidine basic -

Regulation of Calmodulin-Stimulated Cyclic Nucleotide Phosphodiesterase (PDE1): Review
95-105 5/6/06 13:44 Page 95 INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 18: 95-105, 2006 95 Regulation of calmodulin-stimulated cyclic nucleotide phosphodiesterase (PDE1): Review RAJENDRA K. SHARMA, SHANKAR B. DAS, ASHAKUMARY LAKSHMIKUTTYAMMA, PONNIAH SELVAKUMAR and ANURAAG SHRIVASTAV Department of Pathology and Laboratory Medicine, College of Medicine, University of Saskatchewan, Cancer Research Division, Saskatchewan Cancer Agency, 20 Campus Drive, Saskatoon SK S7N 4H4, Canada Received January 16, 2006; Accepted March 13, 2006 Abstract. The response of living cells to change in cell 6. Differential inhibition of PDE1 isozymes and its environment depends on the action of second messenger therapeutic applications molecules. The two second messenger molecules cAMP and 7. Role of proteolysis in regulating PDE1A2 Ca2+ regulate a large number of eukaryotic cellular events. 8. Role of PDE1A1 in ischemic-reperfused heart Calmodulin-stimulated cyclic nucleotide phosphodiesterase 9. Conclusion (PDE1) is one of the key enzymes involved in the complex interaction between cAMP and Ca2+ second messenger systems. Some PDE1 isozymes have similar kinetic and 1. Introduction immunological properties but are differentially regulated by Ca2+ and calmodulin. Accumulating evidence suggests that the A variety of cellular activities are regulated through mech- activity of PDE1 is selectively regulated by cross-talk between anisms controlling the level of cyclic nucleotides. These Ca2+ and cAMP signalling pathways. These isozymes are mechanisms include synthesis, degradation, efflux and seque- also further distinguished by various pharmacological agents. stration of cyclic adenosine 3':5'-monophosphate (cAMP) and We have demonstrated a potentially novel regulation of PDE1 cyclic guanosine 3':5'- monophosphate (cGMP) within the by calpain. -

Expression and Characterization of Pantoea CO Dehydrogenase To
www.nature.com/scientificreports OPEN Expression and characterization of Pantoea CO dehydrogenase to utilize CO-containing industrial Received: 31 October 2016 Accepted: 06 February 2017 waste gas for expanding the Published: 14 March 2017 versatility of CO dehydrogenase Eun Sil Choi1,2,*, Kyoungseon Min3,*, Geun-Joong Kim2, Inchan Kwon1 & Yong Hwan Kim3 Although aerobic CO dehydrogenases (CODHs) might be applicable in various fields, their practical applications have been hampered by low activity and no heterologous expression. We, for the first time, could functionally express recombinant PsCODH in E. coli and obtained a highly concentrated recombinant enzyme using an easy and convenient method. Its electron acceptor spectra, optimum −1 conditions (pH 6.5 and 30 °C), and kinetic parameters (kcat of 12.97 s , Km of 0.065 mM, and specific activity of 0.86 Umg−1) were examined. Blast furnace gas (BFG) containing 20% CO, which is a waste gas from the steel-making process, was tested as a substrate for PsCODH. Even with BFG, the recombinant PsCODH retained 88.2% and 108.4% activity compared with those of pure CO and 20% CO, respectively. The results provide not only a promising strategy to utilize CO-containing industrial waste gases as cheap, abundant, and renewable resources but also significant information for further studies about cascade reactions producing value-added chemicals via CO2 as an intermediate produced by a CODH- based CO-utilization system, which would ultimately expand the versatility of CODH. Carbon monoxide (CO), which is a pollutant in the atmosphere, is massively emitted through both natural (e.g., production by plants and volcanic activity) and artificial processes (e.g., incomplete combustion of fuels and industrial processes)1–3. -

Analyses of PDE-Regulated Phosphoproteomes Reveal Unique and Specific Camp-Signaling Modules in T Cells
Analyses of PDE-regulated phosphoproteomes reveal unique and specific cAMP-signaling modules in T cells Michael-Claude G. Beltejara, Ho-Tak Laua, Martin G. Golkowskia, Shao-En Onga, and Joseph A. Beavoa,1 aDepartment of Pharmacology, University of Washington, Seattle, WA 98195 Contributed by Joseph A. Beavo, May 28, 2017 (sent for review March 10, 2017; reviewed by Paul M. Epstein, Donald H. Maurice, and Kjetil Tasken) Specific functions for different cyclic nucleotide phosphodiester- to bias T-helper polarization toward Th2, Treg, or Th17 pheno- ases (PDEs) have not yet been identified in most cell types. types (13, 14). In a few cases increased cAMP may even potentiate Conventional approaches to study PDE function typically rely on the T-cell activation signal (15), particularly at early stages of measurements of global cAMP, general increases in cAMP- activation. Recent MS-based proteomic studies have been useful dependent protein kinase (PKA), or the activity of exchange in characterizing changes in the phosphoproteome of T cells under protein activated by cAMP (EPAC). Although newer approaches various stimuli such as T-cell receptor stimulation (16), prosta- using subcellularly targeted FRET reporter sensors have helped glandin signaling (17), and oxidative stress (18), so much of the define more compartmentalized regulation of cAMP, PKA, and total Jurkat phosphoproteome is known. Until now, however, no EPAC, they have limited ability to link this regulation to down- information on the regulation of phosphopeptides by PDEs has stream effector molecules and biological functions. To address this been available in these cells. problem, we have begun to use an unbiased mass spectrometry- Inhibitors of cAMP PDEs are useful tools to study PKA/EPAC- based approach coupled with treatment using PDE isozyme- mediated signaling, and selective inhibitors for each of the 11 PDE – selective inhibitors to characterize the phosphoproteomes of the families have been developed (19 21). -

Phosphodiesterase (PDE)
Phosphodiesterase (PDE) Phosphodiesterase (PDE) is any enzyme that breaks a phosphodiester bond. Usually, people speaking of phosphodiesterase are referring to cyclic nucleotide phosphodiesterases, which have great clinical significance and are described below. However, there are many other families of phosphodiesterases, including phospholipases C and D, autotaxin, sphingomyelin phosphodiesterase, DNases, RNases, and restriction endonucleases, as well as numerous less-well-characterized small-molecule phosphodiesterases. The cyclic nucleotide phosphodiesterases comprise a group of enzymes that degrade the phosphodiester bond in the second messenger molecules cAMP and cGMP. They regulate the localization, duration, and amplitude of cyclic nucleotide signaling within subcellular domains. PDEs are therefore important regulators ofsignal transduction mediated by these second messenger molecules. www.MedChemExpress.com 1 Phosphodiesterase (PDE) Inhibitors, Activators & Modulators (+)-Medioresinol Di-O-β-D-glucopyranoside (R)-(-)-Rolipram Cat. No.: HY-N8209 ((R)-Rolipram; (-)-Rolipram) Cat. No.: HY-16900A (+)-Medioresinol Di-O-β-D-glucopyranoside is a (R)-(-)-Rolipram is the R-enantiomer of Rolipram. lignan glucoside with strong inhibitory activity Rolipram is a selective inhibitor of of 3', 5'-cyclic monophosphate (cyclic AMP) phosphodiesterases PDE4 with IC50 of 3 nM, 130 nM phosphodiesterase. and 240 nM for PDE4A, PDE4B, and PDE4D, respectively. Purity: >98% Purity: 99.91% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 1 mg, 5 mg Size: 10 mM × 1 mL, 10 mg, 50 mg (R)-DNMDP (S)-(+)-Rolipram Cat. No.: HY-122751 ((+)-Rolipram; (S)-Rolipram) Cat. No.: HY-B0392 (R)-DNMDP is a potent and selective cancer cell (S)-(+)-Rolipram ((+)-Rolipram) is a cyclic cytotoxic agent. (R)-DNMDP, the R-form of DNMDP, AMP(cAMP)-specific phosphodiesterase (PDE) binds PDE3A directly. -

The Evolution and Population Genetics of the ALDH2 Locus: Random Genetic Drift, Selection, and Low Levels of Recombination
doi: 10.1046/j.1529-8817.2003.00060.x The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination Hiroki Oota1, Andrew J. Pakstis1, Batsheva Bonne-Tamir2, David Goldman3, Elena Grigorenko4, Sylvester L. B. Kajuna5, Nganyirwa J. Karoma5, Selemani Kungulilo6, Ru-Band Lu7, Kunle Odunsi8, Friday Okonofua9, Olga V. Zhukova10, Judith R. Kidd1 and Kenneth K. Kidd1,∗ 1Department of Genetics, Yale University School of Medicine, 333 Cedar Street, P.O. Box 208005, New Haven, CT 06520-8005, USA 2Department of Human Genetics, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel 3Laboratory of Neurogenetics, National Institute of Alcohol Abuse and Alcoholism, Rockville, MD 20852, USA 4Department of Psychology, Yale University, New Haven, CT 06520, USA 5The Hubert Kairuki Memorial University, Dar es Salaam, Tanzania 6Muhimbili University College of Health Sciences, Dar es Salaam, Tanzania 7Department of Psychiatry, Tri-Service General hospital, National Defense Medical Center, Taipei, Taiwan, R.O.C. 8Department of Gynecological Oncology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA 9Department of Obstetrics and Gynecology, Faculty of Medicine, University of Benin, Benin City, Nigeria 10N.I. Vavilov Institute of General Genetics RAS, Moscow, Russia Summary The catalytic deficiency of human aldehyde dehydrogenase 2 (ALDH2) is caused by a nucleotide substitution (G1510A; Glu487Lys) in exon 12 of the ALDH2 locus. This SNP,and four non-coding SNPs, including one in the promoter, span 40 kb of ALDH2; these and one downstream STRP have been tested in 37 worldwide populations. Only four major SNP-defined haplotypes account for almost all chromosomes in all populations. -

How Is Alcohol Metabolized by the Body?
Overview: How Is Alcohol Metabolized by the Body? Samir Zakhari, Ph.D. Alcohol is eliminated from the body by various metabolic mechanisms. The primary enzymes involved are aldehyde dehydrogenase (ALDH), alcohol dehydrogenase (ADH), cytochrome P450 (CYP2E1), and catalase. Variations in the genes for these enzymes have been found to influence alcohol consumption, alcohol-related tissue damage, and alcohol dependence. The consequences of alcohol metabolism include oxygen deficits (i.e., hypoxia) in the liver; interaction between alcohol metabolism byproducts and other cell components, resulting in the formation of harmful compounds (i.e., adducts); formation of highly reactive oxygen-containing molecules (i.e., reactive oxygen species [ROS]) that can damage other cell components; changes in the ratio of NADH to NAD+ (i.e., the cell’s redox state); tissue damage; fetal damage; impairment of other metabolic processes; cancer; and medication interactions. Several issues related to alcohol metabolism require further research. KEY WORDS: Ethanol-to acetaldehyde metabolism; alcohol dehydrogenase (ADH); aldehyde dehydrogenase (ALDH); acetaldehyde; acetate; cytochrome P450 2E1 (CYP2E1); catalase; reactive oxygen species (ROS); blood alcohol concentration (BAC); liver; stomach; brain; fetal alcohol effects; genetics and heredity; ethnic group; hypoxia The alcohol elimination rate varies state of liver cells. Chronic alcohol con- he effects of alcohol (i.e., ethanol) widely (i.e., three-fold) among individ- sumption and alcohol metabolism are on various tissues depend on its uals and is influenced by factors such as strongly linked to several pathological concentration in the blood T chronic alcohol consumption, diet, age, consequences and tissue damage. (blood alcohol concentration [BAC]) smoking, and time of day (Bennion and Understanding the balance of alcohol’s over time. -

Bioalcohol Production by a New Synthetic Route in a Hyperthermophilic Archaeon
COMMENTARY COMMENTARY Bioalcohol production by a new synthetic route in a hyperthermophilic archaeon Volker Müller1 Department of Molecular Microbiology & Bioenergetics, Institute of Molecular Biosciences, Johann Wolfgang Goethe University Frankfurt am Main, 60438 Frankfurt, Germany The still growing demand for bioethanol is establishing an electrochemical ion gradient currently met by two microbiological pro- across the cytoplasmic membrane that then cesses. The by far most important process is drives ATP synthesis by a membrane-bound the use of first- (sugar) or second- (lignocel- ATP synthase (5). One widespread, ferre- lulosic biomass) generation raw materials and doxin-fueled ion-translocating enyzme is + yeast as catalysts. Yeasts convert the sugars the ferredoxin:NAD oxidoreductase found via glycolysis to pyruvate, which is decar- in many bacteria and few archaea (6). In P. furiosus boxylated, and the resulting acetaldehyde is , the reduced ferredoxin is oxidized Fig. 1. Pathways for ethanol formation from glucose in then reduced to ethanol by a monofunctional by a different enzyme, a membrane-bound, yeasts (A) and bacteria (B). Adh1, alcoholdehydrogenase ethanol dehydrogenase. The second and in- multisubunit hydrogenase (Mbh) with evolu- 1; AdhE, bifunctional CoA-dependent ethanol/aldehyde dustrially less important is the bacterial pro- tionarysimilaritytothecomplexIofbacterial dehydrogenase. cess, in which pyruvate is oxidized to acetyl- and mitochondrial electron transport chains CoA, which is then reduced by a bifunctional (7). The Mbh is a proton-reducing (hydrogen aldehyde dehydrogenase/ethanol dehydroge- evolving) and sodium ion-extruding mem- used to drive acetate reduction and not the nase (AdhE) to ethanol. This pathway is brane protein complex (8, 9). generation of a chemiosmotic ion gradient, found in many fermenting bacteria (Fig. -
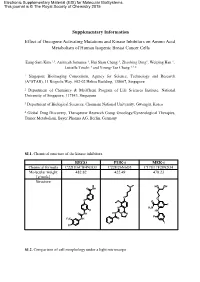
Supplementary Information Effect of Oncogene Activating Mutations And
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2015 Supplementary Information Effect of Oncogene Activating Mutations and Kinase Inhibitors on Amino Acid Metabolism of Human Isogenic Breast Cancer Cells Eung-Sam Kim 1,3, Animesh Samanta 1, Hui Shan Cheng 1, Zhaobing Ding1, Weiping Han 1, Luisella Toschi 4 and Young-Tae Chang 1,2,* 1 Singapore Bioimaging Consortium, Agency for Science, Technology and Research (A*STAR), 11 Biopolis Way, #02-02 Helios Building, 138667, Singapore 2 Department of Chemistry & MedChem Program of Life Sciences Institute, National University of Singapore, 117543, Singapore. 3 Department of Biological Sciences, Chonnam National University, Gwangju, Korea 4 Global Drug Discovery, Therapeutic Research Group Oncology/Gynecological Therapies, Tumor Metabolism, Bayer Pharma AG, Berlin, Germany SI.1. Chemical structure of the kinase inhibitors. REGO PI3K-i MEK-i Chemical formula C22H16ClF4N3O3 C22H26N6O3 C17H17F2IN2O4 Molecular weight 482.82 422.49 478.23 [g/mole] Structure N HO OH O NH O O O O O F N H2N F HN O NH HN N N F F3C NH O N I Cl SI.2. Comparison of cell morphology under a light microscope Scale bar: 100 µm 10x objective lens at Ti microscope (Nikon) SI.3. Scheme of the experimental method for metabolite extraction SI.4. Gradient profile of HPLC run. A : Buffer solution B: Mobile phase H O Time 2 (40 mM NaHPO4, (MeOH/ACN/H2O, Flow rate pH7.8 at RT) 45:45:10, v/v/v) (ml/min) (min) (%) (%) (%) 0 100 0 0 1.0 3 90 10 0 1.0 15 87 13 0 1.0 25 70 30 0 1.0 37 65 35 0 1.0 43 58 42 0 1.0 52 43 57 0 1.0 55 0 100 0 1.0 60 0 0 100 1.0 65 to 80 100 0 0 1.0 SI.5. -
Generate Metabolic Map Poster
Authors: Zheng Zhao, Delft University of Technology Marcel A. van den Broek, Delft University of Technology S. Aljoscha Wahl, Delft University of Technology Wilbert H. Heijne, DSM Biotechnology Center Roel A. Bovenberg, DSM Biotechnology Center Joseph J. Heijnen, Delft University of Technology An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Marco A. van den Berg, DSM Biotechnology Center Peter J.T. Verheijen, Delft University of Technology Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. PchrCyc: Penicillium rubens Wisconsin 54-1255 Cellular Overview Connections between pathways are omitted for legibility. Liang Wu, DSM Biotechnology Center Walter M. van Gulik, Delft University of Technology L-quinate phosphate a sugar a sugar a sugar a sugar multidrug multidrug a dicarboxylate phosphate a proteinogenic 2+ 2+ + met met nicotinate Mg Mg a cation a cation K + L-fucose L-fucose L-quinate L-quinate L-quinate ammonium UDP ammonium ammonium H O pro met amino acid a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar a sugar K oxaloacetate L-carnitine L-carnitine L-carnitine 2 phosphate quinic acid brain-specific hypothetical hypothetical hypothetical hypothetical