Chapter 1 General Introduction
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Y'all Call It Technical and Professional Communication, We Call It
Y’ALL CALL IT TECHNICAL AND PROFESSIONAL COMMUNICATION, WE CALL IT #FORTHECULTURE: THE USE OF AMPLIFICATION RHETORICS IN BLACK COMMUNITIES AND THEIR IMPLICATIONS FOR TECHNICAL AND PROFESSIONAL COMMUNICATION STUDIES by Temptaous T. Mckoy July 2019 Director of Dissertation: Michelle F. Eble Major Department: Department of English This project seeks to define and identify the use of Amplification Rhetorics (AR) in the social movement organization TRAP Karaoke and at three Historically Black Colleges and Universities (HBCUs). In highlighting the AR practices in Black communities. This project will provide insight into what these practices and spaces have to offer to the field of Technical and Professional Communication (TPC). AR is a theoretical framework that details the discursive and communicative practices, both written/textual and embodied/performative performed/used by individuals that self-identify as Black and centers the lived experiences and epistemologies of Black people and other historically marginalized groups. AR are #ForTheCulture practices that provoke change within the academy, regardless the field, however for this project I focus on the field of TPC. The phrase “For the Culture,” is used to describe or allude to an action that can benefit those individually or within a community much like TPC artifacts. This project identifies rhetorical practices located in Black spaces and communities that can be identified as TPC through the reclamation of agency, the sharing of narratives, and the inclusion of Black epistemologies. It illustrates just what it means to pass the mic and remind folks that we not ‘bout to act like there aren’t people of color at the TPC table. -
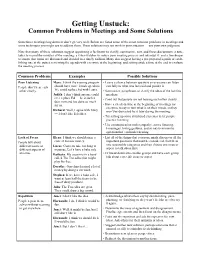
Getting Unstuck: Common Problems in Meetings and Some Solutions
Getting Unstuck: Common Problems in Meetings and Some Solutions Sometimes meetings bog down or don’t go very well. Below are listed some of the most common problems in meetings and some techniques you might use to address them. These solutions may not work in your situation — use your own judgment. Note that many of these solutions suggest appointing a facilitator to clarify, summarize, sort, and focus discussions; a note- taker to record the minutes of the meeting; a vibes-watcher to notice poor meeting process and interrupt it; and a timekeeper to ensure that items are discussed and decided in a timely fashion. Many also suggest having a pre-prepared agenda or estab- lishing one at the outset, reviewing the agenda with everyone at the beginning, and setting aside a time at the end to evaluate the meeting process. Common Problems Examples Possible Solutions Poor Listening Mary: I think the evening program • Leave a silence between speakers so everyone can listen People don’t hear each should have more female speakers. carefully to what was last said and ponder it. other clearly. We could replace Ed with Laura. • Summarize, paraphrase, or clarify the ideas of the last few Judith: I don’t think anyone could speakers. ever replace Ed — he is such a • Point out that people are not hearing each other clearly. dear man and has done so much for us. • Have a check-in time at the beginning of meetings for everyone to say in turn what is on their minds, so they Richard: Well, I agree with Mary won’t be distracted by it later during the meeting. -

Dj Khaled 2011 Album Tracklist
Dj khaled 2011 album tracklist Khaled took to Twitter to debut his new album cover on which he chooses to sit like a boss. The project is set for a June 28th release date, DJ Khaled – We The Best Forever (Album Cover & Track List) June 27, K Views. We the Best Forever is the fifth studio album by DJ Khaled. It was released under We the Best 4 Track listing; 5 Charts. Weekly In February Khaled confirmed that Drake, Rick Ross, T-Pain, and Plies will be featured in the album.Background · Singles · Reception · Track listing. Title, Album details, Peak chart positions, Certifications. US Released: July 19, ; Label: We the Best, Young Money, Cash Singles · Guest appearances · Music videos · Production credits. By Alvin Blanco June 27, PM That said, the tracklist for DJ Khaled's fifth album, 'We the Best Forever,' has now arrived, and it's a star-studded affair. From DJ Khaled's second album, We the Best, this song features Akon, .. DJ Khaled's sixth studio album, has a couple of good tracks, this. June 27, · Posted in news, tracklist · 18 Comments · dj-khaled-we-the-best-forever-album-cover. Amazon just revealed the The album drops July 19th. DJ Khaled. Released July 19, We the Best Forever is the fifth studio album by DJ Khaled, released on July 19, through Cash Money Records. DJ Khaled new songs, albums, biography, chart history, photos, videos, news, and more on Billboard, the go-to source for what's hot in music. Tracklist with lyrics of the album WE GLOBAL [] from DJ Khaled: This album was submitted on March 3rd, and last modified on July 25th, Release: , Album: DJ Khaled - We the Best Forever; Tracklist: 1 I'm On One (feat. -

Transcript of Dr. Pamela Shipp ’69 Interviewed on May 2, 2019 by Kirk Woundy
Transcript of Dr. Pamela Shipp ’69 Interviewed on May 2, 2019 by Kirk Woundy Kirk Woundy: This is an interview with Dr. Pamela Shipp for Untold Stories, May 2, 2019. We’ll start by talking about your childhood a little bit. Dr. Pamela Shipp ’69: Ooh, I love talking about my childhood. I was just really a blessed child. I grew up in East St. Louis. I was born in St. Louis and our family ... Well, then we lived in East St. Louis, in a very large family. My mom’s the youngest of 10 kids, really 11 but one died, so she says 10, and we go with 10, and hopefully ... So, one of the good things about growing up in a large family, for me, I always felt loved, and I always felt that I belonged, and it was just absolutely refreshing and sometimes comical because when we moved ... PS: I moved to Denver with my mom who took a job in Colorado and we moved to Denver where I discovered that everyone didn’t love me, and of course, I’m just a little kid but still I’m anticipating that everyone is going to want me and love me and be there for me, which was not the case when we moved and I noticed that in elementary school. So that was a nice kind of ... I think it was a nice understanding that I got to have very early that I still had the foundation of knowing that I was loved and lovable but knowing that there are people that are not going to love me, and I learned that at an early age, so that’s why I think that was kind of good for me. -

I Still Am Album Zip Download Torrents Download Various Artists - Pop Hits in Reggae Playlist (2010) Album
i still am album zip download torrents Download Various Artists - Pop Hits in Reggae Playlist (2010) Album. 1. I Am Still in Love 2. I Don't Want to Talk About It 3. The Way You Do the Things You Do 4. It's All in the Game 5. My Sweet Lord 6. Baltimore 7. I Want to Hold Your Hand 8. Natural Mystic 9. Ain't No Sunshine 10. Stand by Your Man 11. Unexpected Places 12. Let It Be 13. O.K Fred (My Name Is Fred) 14. The Further You Look 15. Never Never 16. Hey Jude 17. You Will Never Find 18. Wear You to the Ball 19. Stick by Me 20. The Tide Is High 21. Puppet on a String 22. Waiting in Vain 23. By the Rivers of Babylon 24. Crazy Bald Head 25. Midnight Cowboy 26. No Woman No Cry 27. In the Summertime 28. Help Me Make It Through the Night 29. No Satisfaction 30. Kingston Town 31. Back for Good 32. Summertime 33. Wet Dream 34. My Baby 35. Sweet Sensation 36. Angel of the Morning 37. Love Me Tender 38. Liquidator Dub 39. Red Red Wine 40. Lonely Girl 41. Cherry Oh Baby 42. Mona Lisa 43. Imagine 44. So Much Trouble 45. A House Is Not a Home 46. You're Always on My Mind 47. Living in the Footsteps 48. Train to Skaville 49. Hold Me Tight. DOWNLOAD ALBUM: Roddy Ricch – Please Excuse Me For Being Antisocial [Zip File] Roddy Ricch released this music ALBUM in 2019 titled Please Excuse Me For Being Antisocial and its available for easy download and streaming. -

Students' Book
หนังสือเรียน รายวิชาพื้นฐาน ภาษาอังกฤษ STUDENTS’ BOOK ชั้นมัธยมศึกษาปีที่ 6 กลุ่มสาระการเรียนรู้ภาษาต่างประเทศ ตามหลักสูตรแกนกลางการศึกษาขั้นพื้นฐาน พุทธศักราช 2551 ผู้เรียบเรียง ผู้ตรวจ บรรณาธิการ Sue Kay ดร.ไพสาร บุญประกอบ นางสาววราภรณ์ เลิศวรสิริกุล Vaughan Jones นางสาววารี สวนสุวรรณ นางสาวจุฑาวรรณ ศรีศิริโชคชัย Daniel Brayshaw นางสุจิตรา สัตยารักษ์ นางหรรษา วงศ์กิดาการ พิมพ์ครั้งที่ 1 พุทธศักราช 2562 จำานวน 45,000 เล่ม ISBN 978-616-559-053-2 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0 p. 4 Grammar: Present tenses – p. 5 Grammar: Quantifiers p. 6 Grammar: Present Perfect and p. 7 Grammar: p. 8 Grammar: Future p. 9 Grammar: First and p. 10 Grammar: Modal p. 11 Grammar: Defining review Vocabulary: Food and drink Past Simple Comparative and forms Second Conditionals verbs for obligation and relative clauses Vocabulary: Houses Vocabulary: Shops superlative adjectives; too Vocabulary: Books and Vocabulary: Technology permission Vocabulary: Work and enough films and the Internet Vocabulary: Education Vocabulary: Clothes and accessories 1 Vocabulary Grammar Listening Reading Grammar Speaking Writing Looks pp. 12–13 Clothes and accessories; p. 14 Dynamic and state verbs p. 15 Friendship pp. 16–17 Genes p. 18 Present Perfect p. 19 Talking about a photo pp. 20–21 A description of a person Verb phrases to do with clothes; Exam Focus: Multiple choice Exam Focus: Multiple Continuous Vocabulary: adjective Synonyms – appearance and Pronunciation: Numbers choice order personality Vocabulary: Relationship phrases Vocabulary: Compound Reading: Facebook profile photos adjectives – appearance and what they mean pp. 24–25 Sport; Compound p. 26 Narrative tenses p. 27 Role models pp. 28–29 A Paralympic p. 30 Verb patterns p. 31 Asking for and giving pp. 32–33 An article 2 nouns; Sport collocations; People Exam Focus: Sentence completion athlete an opinion; Agreeing and in sport Pronunciation: Long vowel sounds Exam Focus: Gapped text disagreeing Just do it! Reading and listening: Sports quiz Vocabulary: Word families – Vocabulary: Phrasal verbs personal qualities pp. -

If You're an Avid Hip-Hop Fan, Surely
Coalition Of (916) Magazine ACE Magazine Independent Crunk Magazine Da Streetz Magazine Sacramento, CA West Palm Beach, FL Atlanta, GA New York, NY www.the916.com 772-332-7979 www.crunkmag.com www.dastreetzonline.com 916-452-2482 News 404-756-9868 212-589-1009 Don Diva Magazine Exposure Magazine F.E.D.S. Magazine Tha Hole Magazine Holla Magazine Kronick Magazine New York, NY Chicago, IL New York, NY Huntsville, AL Atlanta, GA Los Angeles, CA 877-366-3482 847-366-8915 1369 Madison Ave #406 256-652-0804 www.hollamag.com www.kronick.com www.exposuremagazines.com NY, NY 10128 www.thahole.com Murder Dog Magazine MVP Magazine New Power Magazine Next Level Magazine OZONE Magazine Rude Magazine Los Angeles, CA Jacksonville, FL Columbus, MS New York, NY Orlando, FL New Orleans, LA www.murderdog.com www.mvpthemag.com 662-251-0075 thenextlevelmagazine.com 407-447-6063 504-246-1491 707-553-1850 904-493-6075 newpowermagazine.com www.ozonemag.com Showcase Magazine Street Masters Magazine Streetz Magazine Urban Pages Magazine UrbLife Magazine Xplosive Magazine Los Angeles, CA Memphis, TN Virginia Charleston, SC Montgomery, AL San Francisco, CA 510-481-3080 901-888-1003 800-770-1078 [email protected] www.urblife.com 925-427-9330 843-747-5131 888-683-0677 [email protected] Check out other magazines like ours online at www.MAGCOALITION.com PUBLISHERS: Julia Beverly (JB) Chino EDITOR-IN-CHIEF: Julia Beverly (JB) ASSISTANT EDITOR: Noel Malcolm MUSIC EDITORS: ADG & CL Mecca CONTRIBUTORS: AJ Woodson, Big Earl, Bogan, Brian O’Hare, Charlie Braxton, Chris Imani, Cynthia Coutard, Dain Burroughs, Darnella Dunham, Dave Goodson, Felita Knight, Hasan Brown, Iisha Hillmon, Jeska Manrique, Jesse Jazz, Jessica Koslow, J Lash, Katerina Perez, Keith Kennedy, K.G. -

November 1942–February 1943
Chapter 3 November 1942–February 1943 November 1, 1942 Dear Ray, I received your letter and also the check. I sure do appreciate the money and I will pay you every cent when I come back from the furlough. I am trying to get it at the end of this month but they haven’t told me if it was ok yet. They are letting twenty-five at a time go. The first bunch came back today and the second twenty- five leave tomorrow. I will probably be one of the third twenty-five, I hope. I told Mom I was coming, Ray, so she ought to be glad. She was always asking when they were going to give me a furlough and now they are going to. The way I have it figured out, it will take me about four days to get home, stay about seven and four days back. That will be my full fifteen days. I received the magazine you sent and I must say you look pretty good alongside the bishop and with that trophy, too. Boy, you must be doing an awful lot for those kids in your parish. I hope they appreciate all that you are doing for them. Eddie training in the desert–October 1942. 1 Well, Ray, we are getting ready to leave the desert. They tell us we will be sent out of here by the fifteenth of this month and then back to Camp Cooke. It sure will be great to get back to camp. I am having some pictures developed and I will bring them home or mail them to either you or home. -

Featuring Lil Baby Today Performing on the To
FOR IMMEDIATE RELEASE YO GOTTI RELEASES NEW MUSIC VIDEO “PUT A DATE ON IT” FEATURING LIL BABY TODAY PERFORMING ON THE TONIGHT SHOW STARRING JIMMY FALLON JANUARY 29 TENTH FULL-LENGTH ALBUM COMING SOON (January 25, 2019 – Los Angeles, CA) – Today, multiplatinum Memphis superstar rapper and CMG mogul Yo Gotti returns with a brand new single entitled “Put A Date On It” [feat. Lil BAby] today. Get it HERE via CMG/Epic Records. Once again, the hip-hop titan turns up with a hard-hitting and hypnotic anthem that highlights his verbal elasticity and propensity for hashtaggable hooks. Meanwhile, Atlanta’s Lil BAby delivers a show-stopping cameo of his own, capitalizing on undeniable chemistry with his Memphis counterpart. Additionally, Gotti shares the official music video. Continuing a tradition of larger-than-life clips, he hosts a Last Supper-style dinner not just in the hood, but literally in the middle of the streets! Invited to this O.G. gathering are none other than CMG heavyweights MoneybAgg Yo, Blocboy JB, and BlAc YoungstA. View the clip on VEVO/YouTube HERE! Next up, he performs the song for the first time on late-night television when he graces the stage at The Tonight Show Starring Jimmy Fallon on January 29. Be sure to tune in. “Put A Date On It” kicks off what promises to be a landmark 2019 for Gotti. It serves as the single and first taste of his tenth full-length—arriving very soon. It also follows up a busy 2018, which saw him team up with G- Eazy for the banger “1942” (over 101 million Spotify streams to date) and T.I. -

Yo Gotti's Highly Anticipated New Album Untrapped Out
YO GOTTI’S HIGHLY ANTICIPATED NEW ALBUM UNTRAPPED OUT NOW NEW MUSIC VIDEO “POSE” FEATURING MEGAN THEE STALLION & LIL UZI VERT LIVE AT 12PM ET INCLUDES THE SINGLES “H.O.E. (HEAVEN ON EARTH)” & “MORE READY THAN EVER” HOSTS HIGH-PROFILE ALBUM LAUNCH DINNER AT FOUR SEASONS (January 31, 2020 – Los Angeles, CA) – After maximizing anticipation throughout the past month, multiplatinum Memphis superstar rapper and CMG mogul Yo Gotti unveils his anxiously awaited tenth full-length studio album and first in three years, Untrapped, today. Upon going live, it catapulted into the Top 2 of iTunes Top Hip-Hop/Rap Albums Chart where it still remains. Get it HERE via Inevitable/Epic Records. To celebrate the release, he hosted a live chat last night online and will be debuting the new “Pose” music video at 12pm EST. Watch it HERE. On Tuesday, he welcomed a bevy of tastemakers, DJs, on-air personalities, celebrities, and peers to experience Untrapped before anyone during a posh private dinner at the Four Seasons in New York City. Rather than ban smartphones, the MC encouraged social interaction, posts, and tweets. As a result, social media lit up with buzz for the impending release and heightened excitement. He also preceded the record with single “H.O.E. (Heaven On Earth).” In under a week, it tallied 1.3 million Spotify streams and 1 million-plus views on the music video. Straight out of the gate, it garnered major looks from Billboard and more as Hypebeast promised, “Yo Gotti has been cooking up something major in the past two years.” Earlier this week, he also pulled up on Funkmaster Flex for what has become a much-talked about Freestyle. -
Ex College Dropout Elected A.S. President
Volume 146 • Issue 39 Serving San Jose State University since 1934 Th ursday, May 5, 2016 SJSUNEWS.COM/SPARTAN_DAILY VIDEOS AT YOUTUBE.COM/C/SPARTANDAILYSJSU Americans need to end the OPINION P5: murdering of innocent babies SPORTS P7: San Jose State holds on to even the season series against the Lopes CLIMBING TO THE TOP HEADS UP Ex college dropout elected A.S. president By Itzel Castro Contributing writer Th ere’s nothing like a working in blue-collar brush with death to get jobs, Perea decided to you back on track. Th at’s give college a second part of the journey that chance and returned to Hector Perea has taken. Fresno City College in Perea, the new A.S. Fall 2012 and ultimately President for Fall graduated, transferring 2016, was involved in a to San Jose State gruesome car accident University in Fall 2014. that nearly took his life Now, as a 28-year- in his early 20s. old SJSU student, the Kavin Mistry | Spartan Daily He was a community San Jose native son of Members of SJSU Cru, Rolan Jimenez (left), Gabe Senbetu (center left), Luke Bryant (center right) and college dropout Mexican immigrants Zack Burgess (right) play spike ball at Tower Lawn. working a 9-to-5 job as looks back, mostly with a construction worker pride, on his early days. CINCO DE MAYO in West Virginia, where “My setbacks in life he almost faced death are my biggest learning aft er an exhausting day experiences because of work. I was never the best Take a little trip in a lowrider “Af ter that car student and never By Rashaad Mubarak accident, I had a life- appreciated education, Copy editor changing epiphany, but experiencing all where I realized I had those low points in my to go back to school,” life made me want to Some Americans assume Th is subculture started Perea said. -

Ray J Sean Paul
YOUR FAVORITE RAPPER’S FAVORITE MAGAZINE OZONE MAGAZINE WE WORK NIGHTS,WE SOMEVAMPIRES / GATHER ROUNDTHE BEAT LIKE A CAMPFIRE ISSUE #78 NEW BOYZ RAY J SHORTY MACK ACE MAINO HOOD SEAN YO GOTTI PAUL RICH JW KIDS WILLY JAVON NORTHPOLE BLACK LOLA LUV UNLADYLIKE 1 OZONE MAG // YOURYOUR FAVORITEFAVORITE RAPPER’SRAPPER’S FAVORITEFAVORITE MAGAZINEMAGAZINE KNOCKOKNOCKOUUTT ENTERTAINMENT’sENTERTAINMENT’s SHORTYSHORTY MACKMACK && RAYRAY JJ ACE HOOD MAINO NEW BOYZ SEAN PAUL YO GOTTI RICH KIDS WILL Y JAVON BLACK NORTHPOLE UNLADYLIKE LOLA LUV 20 // OZONE MAG 2 // OZONE MAG OZONE MAG // 3 4 // OZONE MAG OZONE MAG // 5 6 // OZONE MAG OZONE MAG // 7 8 // OZONE MAG OZONE MAG // 9 PUBLISHER/EDITOR-IN-CHIEF // Julia Beverly MUSIC EDITOR // Randy Roper FEATURES EDITOR // Eric N. Perrin cover stories ASSOCIATE EDITOR // Maurice G. Garland GRAPHIC DESIGNER // David KA 42-43 ACE HOOD ADVERTISING SALES // Che Johnson, Gary Archer 54-58 RAY J & SHORTY MACK PROMOTIONS DIRECTOR // Malik Abdul W12-W13 WILLY NORTHPOLE SPECIAL EDITION EDITOR // Jen McKinnon WEST COAST EDITOR-AT-LARGE // D-Ray LEGAL CONSULTANT // Kyle P. King, P.A. SUBSCRIPTIONS MANAGER // Adero Dawson ADMINISTRATIVE // Kisha Smith INTERNS // Devon Buckner, Jee’Van Brown, Krystal Moody, Memory Martin, Ms Ja, Shanice Jarmon, Torrey Holmes CONTRIBUTORS // Anthony Roberts, Bogan, Camilo Smith, Charlamagne the God, Chuck T, Cierra Middlebrooks, David Rosario, Diwang Valdez, DJ BackSide, Edward Hall, E-Z Cutt, Gary Archer, Hannibal Matthews, Jacquie Holmes, J Lash, Jason Cordes, Jelani Harper, Joey Colombo, Johnny Louis, Kay Newell, Keadron Smith, Keita Jones, Keith Kennedy, K.G. Mosley, King Yella, Luis Santana, Luvva J, Luxury Mindz, Marcus DeWayne, Matt Sonzala, Maurice G.