Redalyc.Microorganisms Screening for Limonene Oxidation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biosynthesis of New Alpha-Bisabolol Derivatives Through a Synthetic Biology Approach Arthur Sarrade-Loucheur
Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach Arthur Sarrade-Loucheur To cite this version: Arthur Sarrade-Loucheur. Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach. Biochemistry, Molecular Biology. INSA de Toulouse, 2020. English. NNT : 2020ISAT0003. tel-02976811 HAL Id: tel-02976811 https://tel.archives-ouvertes.fr/tel-02976811 Submitted on 23 Oct 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE En vue de l’obtention du DOCTORAT DE L’UNIVERSITÉ DE TOULOUSE Délivré par l'Institut National des Sciences Appliquées de Toulouse Présentée et soutenue par Arthur SARRADE-LOUCHEUR Le 30 juin 2020 Biosynthèse de nouveaux dérivés de l'α-bisabolol par une approche de biologie synthèse Ecole doctorale : SEVAB - Sciences Ecologiques, Vétérinaires, Agronomiques et Bioingenieries Spécialité : Ingénieries microbienne et enzymatique Unité de recherche : TBI - Toulouse Biotechnology Institute, Bio & Chemical Engineering Thèse dirigée par Gilles TRUAN et Magali REMAUD-SIMEON Jury -

Investigating Labdane-Related Diterpenoids Biosynthetic Network In
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2012 Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase Yisheng Wu Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry Commons Recommended Citation Wu, Yisheng, "Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase" (2012). Graduate Theses and Dissertations. 12680. https://lib.dr.iastate.edu/etd/12680 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Investigating labdane-related diterpenoids biosynthetic network in rice: Characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase by Yisheng Wu A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Reuben Peters, Major Professor Gustavo MacIntosh Thomas Bobik Bing Yang Mark Hargrove Iowa State University Ames, Iowa 2012 Copyright © Yisheng Wu , 2012. All -

Pea SAD Short-Chain Dehydrogenase/Reductase Author to Whom All Correspondence Should Be Addressed
Plant Physiology Preview. Published on February 22, 2011, as DOI:10.1104/pp.111.173336 1 Running head: Pea SAD Short-Chain Dehydrogenase/Reductase Author to whom all correspondence should be addressed: Åke Strid Akademin för naturvetenskap och teknik och Centrum för livsvetenskap Örebro universitet SE-70182 Örebro Sweden Telephone: +46-19-303603 E-mail: [email protected] Most appropriate journal research area: Cell Biology 1 Copyright 2011 by the American Society of Plant Biologists 2 The Pisum sativum SAD Short-Chain Dehydrogenase/Reductase: Quinone Reduction, Tissue 1 Distribution, and Heterologous Expression Nikolai Scherbak2, Anneli Ala-Häivälä2, Mikael Brosché2, Nathalie Böwer, Hilja Strid, John R. Gittins3, Elin Grahn, Leif A. Eriksson4, and Åke Strid* Akademin för naturvetenskap och teknik och Centrum för livsvetenskap (N.S., A.A.-H., N.B., E.G., L.A.E., Å.S) and Hälsoakademin och Centrum för livsvetenskap (H.S.), Örebro universitet, S-70182 Örebro, Sweden; Biokemi och biofysik (M.B., J.R.G.), Institutionen för Kemi, Göteborgs universitet, P.O. Box 462, S-405 30 Göteborg, Sweden; Division of Plant Biology (M.B.), Department of Biosciences, University of Helsinki, P.O. Box 65, FIN-00014 Helsinki, Finland, and Institute of Technology, University of Tartu, Nooruse 1, Tartu 50411, Estonia. 2 3 Footnotes: 1This work was supported by Helge Ax:son Johnson’s foundation (N.S.), the Carl Trygger Foundation (Å.S.) and the Faculty for Science and Technology at Örebro University (Å.S.). The Strategic Network for Swedish Plant Biotechnology (SSF) funded J.R.G.’s stay in Sweden with a grant to Å.S. -
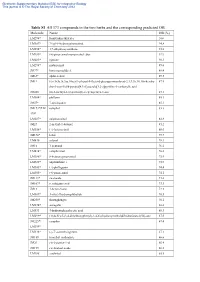
Table S1 All 373 Compounds in the Two Herbs and the Corresponding
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Table S1 All 373 compounds in the two herbs and the corresponding predicted OB Molecule Name OB (%) LM294* forsythidmethylester 100 LM387* 7'-epi-8-hydroxypinoresinol 94.8 LM394* 1,7-dihydroxyxanthone 93.8 LM289* (+)-pinoresinol monomethyl ether 92.9 LM285* egenine 90.3 LM298* matairesinol 89.6 JM77* loniceracetalide A 89.4 JM63* alpha-cedrol 89.3 JM1* (-)-(3r,8s,9r,9as,10as)-9-ethenyl-8-(beta-d-glucopyranosyloxy)-2,3,9,9a,10,10a-hexahy 87.5 dro-5-oxo-5h,8h-pyrano[4,3-d]oxazolo[3,2-a]pyridine-3-carboxylic acid JM200 (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one 87.1 LM304* phillyrin 85.1 JM57* 7-epi-loganin 85.1 JM173*/LM camphol 83.3 398* LM287* epipinoresinol 82.8 JM27 2-methyl-1-butanol 81.2 LM356* (+)-lariciresinol 80.0 JM170* ledol 79.7 LM430 safynol 78.3 JM16 1-pentanol 76.2 LM414* campherenol 76.0 LM386* 8-hydroxypinoresinol 75.9 LM428* onjixanthone i 75.9 LM345* (+)-phillygenin 74.4 LM305* (+)-pinoresinol 74.1 JM113* sweroside 73.6 JM107* secologanic acid 73.3 JM13 1-hexen-3-one 72.3 LM309* 3-ethyl-7hydroxyphthalide 70.5 JM250* shuangkangsu 70.1 LM274* astragalin 68.6 LM331 4-hydroxyphenylacetic acid 68.3 LM299* (3r,4s,5r)-5-(3,4-dimethoxyphenyl)-3,4-bis(hydroxymethyl)dihydrofuran-2(3h)-one 67.5 JM225*/ camphor 67.4 LM399* LM338* (-)-7’-o-methylegenine 67.1 JM189 trimethyl isothiazole 66.6 JM29 cis-2-penten-1-ol 66.4 JM199 cis-linalool oxide 66.2 LM288 erythritol 65.8 Electronic Supplementary Material (ESI) for -

NPC Natural Product Communications
NPC-CMAPSEEC Issue Volume 12. Issue 2. Pages 153-318. 2017 ISSN 1934-578X (printed); ISSN 1555-9475 (online) www.naturalproduct.us NPC Natural Product Communications EDITOR-IN-CHIEF HONORARY EDITOR DR. PAWAN K AGRAWAL PROFESSOR GERALD BLUNDEN Natural Product Inc. The School of Pharmacy & Biomedical Sciences, 7963, Anderson Park Lane, University of Portsmouth, Westerville, Ohio 43081, USA Portsmouth, PO1 2DT U.K. [email protected] [email protected] EDITORS ADVISORY BOARD PROFESSOR ALEJANDRO F. BARRERO Department of Organic Chemistry, University of Granada, Prof. Giovanni Appendino Prof. Niel A. Koorbanally Campus de Fuente Nueva, s/n, 18071, Granada, Spain Novara, Italy Durban, South Africa [email protected] Prof. Norbert Arnold Prof. Chiaki Kuroda PROFESSOR MAURIZIO BRUNO Halle, Germany Tokyo, Japan Department STEBICEF, Prof. Hartmut Laatsch University of Palermo, Viale delle Scienze, Prof. Yoshinori Asakawa Gottingen, Germany Parco d’Orleans II - 90128 Palermo, Italy Tokushima, Japan [email protected] Prof. Marie Lacaille-Dubois Prof. Vassaya Bankova PROFESSOR VLADIMIR I. KALININ Dijon, France Sofia, Bulgaria G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Prof. Shoei-Sheng Lee Far Eastern Branch, Russian Academy of Sciences, Prof. Roberto G. S. Berlinck Taipei, Taiwan Pr. 100-letya Vladivostoka 159, 690022, São Carlos, Brazil Vladivostok, Russian Federation Prof. Anna R. Bilia Prof. M. Soledade C. Pedras [email protected] Saskatoon, Canada Florence, Italy PROFESSOR YOSHIHIRO MIMAKI Prof. Geoffrey Cordell Prof. Luc Pieters School of Pharmacy, Antwerp, Belgium Chicago, IL, USA Tokyo University of Pharmacy and Life Sciences, Prof. Fatih Demirci Prof. Peter Proksch Horinouchi 1432-1, Hachioji, Tokyo 192-0392, Japan Düsseldorf, Germany [email protected] Eskişehir, Turkey Prof. -

Monoterpene Biosynthesis in the Glandulartrichomes Of
Plant Physiol. (1989) 89, 1351-1357 Received for publication July 1, 1988 0032-0889/0000/1351 /07/$01 .00/0 and in revised form December 2, 1988 Biochemical and Histochemical Localization of Monoterpene Biosynthesis in the Glandular Trichomes of Spearmint (Mentha spicata)" 2 Jonathan Gershenzon, Massimo Maffei3, and Rodney Croteau* Institute of Biological Chemistry, Washington State University, Pullman, Washington 99164-6340 ABSTRACT type diterpenes found in the glandular exudate. The head cells The primary monoterpene accumulated in the glandular tri- oftobacco trichomes are the only leafcells able to incorporate chomes of spearmint (Mentha spicata) is the ketone (-)-carvone radiolabeled acetate into duvane diterpenes; removal ofthese which is formed by cyclization of the C10 isoprenoid intermediate cells severely reduces or eliminates duvane production (22). geranyl pyrophosphate to the olefin (-)-limonene, hydroxylation Thus, it is tempting to speculate that all terpenes found in to (-)-trans-carveol and subsequent dehydrogenation. Selective glandular trichomes are synthesized in situ. However, recent extraction of the contents of the glandular trichomes indicated reports of monoterpene synthesis by undifferentiated cells in that essentially all of the cyclase and hydroxylase activities culture (5, 28), coupled to evidence for monoterpene transport resided in these structures, whereas only about 30% of the in intact plants (7, 31), suggest that the site of synthesis may carveol dehydrogenase was located here with the remainder not necessarily be the same as the site of accumulation. located in the rest of the leaf. This distribution of carveol dehy- drogenase activity was confirmed by histochemical methods. Spearmint (Mentha spicata, Lamiaceae) accumulates large Electrophoretic analysis of the partially purified carveol dehydro- quantities of monoterpenes in glandular trichomes, the major genase from extracts of both the glands and the leaves following constituent of which is (-)-carvone (20). -

Frankincense Essential Oil Prepared from Hydrodistillation of Boswellia
Ni et al. BMC Complementary and Alternative Medicine 2012, 12:253 http://www.biomedcentral.com/1472-6882/12/253 RESEARCH ARTICLE Open Access Frankincense essential oil prepared from hydrodistillation of Boswellia sacra gum resins induces human pancreatic cancer cell death in cultures and in a xenograft murine model Xiao Ni1, Mahmoud M Suhail2, Qing Yang3, Amy Cao4, Kar-Ming Fung3,5,6, Russell G Postier7, Cole Woolley8, Gary Young8, Jingzhe Zhang1* and Hsueh-Kung Lin3* Abstract Background: Regardless of the availability of therapeutic options, the overall 5-year survival for patients diagnosed with pancreatic cancer remains less than 5%. Gum resins from Boswellia species, also known as frankincense, have been used as a major ingredient in Ayurvedic and Chinese medicine to treat a variety of health-related conditions. Both frankincense chemical extracts and essential oil prepared from Boswellia species gum resins exhibit anti-neoplastic activity, and have been investigated as potential anti-cancer agents. The goals of this study are to identify optimal condition for preparing frankincense essential oil that possesses potent anti-tumor activity, and to evaluate the activity in both cultured human pancreatic cancer cells and a xenograft mouse cancer model. Methods: Boswellia sacra gum resins were hydrodistilled at 78°C; and essential oil distillate fractions were collected at different durations (Fraction I at 0–2 h, Fraction II at 8–10 h, and Fraction III at 11–12 h). Hydrodistillation of the second half of gum resins was performed at 100°C; and distillate was collected at 11–12 h (Fraction IV). Chemical compositions were identified by gas chromatography–mass spectrometry (GC-MS); and total boswellic acids contents were quantified by high-performance liquid chromatography (HPLC). -

Supplementary Information
Supplementary Information The domain composition of sequences from Laurencia dendroidea that were annotated as terpene synthases and presented in Table 2 of the main manuscript were obtained through search for conserved domains using the NCBI Conserved Domain Database (CDD, [1]) and compared with the domain composition of corresponding sequences available in the SwissProt/UniProt and PlantCycDB databases. Analyzing the tables and figures below, we reinforce the Blast annotations presented in Table 2 of the main manuscript text, although biochemical and gene cloning approaches are necessary to prove these in silico identifications. For two of the 21 terpene synthase sequences from Laurencia (identified as nerolidol synthase and (+)-delta-cadinene synthase), we were not able to identify conserved domains, possibly because they were partial sequences. In the case of alpha-bisabolene synthase, we provide the domain composition of a reference sequence and the alignment of the sequence from L. dendroidea with this reference, showing a high similarity between them. Table S1. List of domain hits for a putative (3R)-linalool synthase gene from L. dendroidea. Name Accession Description Interval E-Value DNA repair protein (rad1); All proteins in this rad1 TIGR00596 1–51 1.04e-15 family for which functions are known are ... Figure S1. Conserved domains detected in a putative (3R)-linalool synthase gene from L. dendroidea (edited from the NCBI CDD database, [1]). Table S2. List of domain hits for the (3R)-linalool synthase gene from Zea mays mays (GDQC-116173-MONOMER—PlantCycDB). Name Accession Description Interval E-Value Plant Terpene Cyclases, Class 1; Terpene_cyclase_plant_C1 cd00684 This CD includes a diverse group of 1377–1833 9.57e-107 monomeric plant terpene .. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

Mycofactocin-Associated Mycobacterial Dehydrogenases with Non-Exchangeable NAD Cofactors
www.nature.com/scientificreports OPEN Mycofactocin-associated mycobacterial dehydrogenases with non-exchangeable NAD Received: 14 October 2016 Accepted: 12 December 2016 cofactors Published: 25 January 2017 Daniel H. Haft1, Phillip G. Pierce2,3, Stephen J. Mayclin2,3, Amy Sullivan2,3, Anna S. Gardberg2,3, Jan Abendroth2,3, Darren W. Begley2,3,†, Isabelle Q. Phan2,4, Bart L. Staker2,4, Peter J. Myler2,4,5, Vasilios M. Marathias2,3, Donald D. Lorimer2,3 & Thomas E. Edwards2,3 During human infection, Mycobacterium tuberculosis (Mtb) survives the normally bacteriocidal phagosome of macrophages. Mtb and related species may be able to combat this harsh acidic environment which contains reactive oxygen species due to the mycobacterial genomes encoding a large number of dehydrogenases. Typically, dehydrogenase cofactor binding sites are open to solvent, which allows NAD/NADH exchange to support multiple turnover. Interestingly, mycobacterial short chain dehydrogenases/reductases (SDRs) within family TIGR03971 contain an insertion at the NAD binding site. Here we present crystal structures of 9 mycobacterial SDRs in which the insertion buries the NAD cofactor except for a small portion of the nicotinamide ring. Line broadening and STD-NMR experiments did not show NAD or NADH exchange on the NMR timescale. STD-NMR demonstrated binding of the potential substrate carveol, the potential product carvone, the inhibitor tricyclazol, and an external redox partner 2,6-dichloroindophenol (DCIP). Therefore, these SDRs appear to contain a non-exchangeable NAD cofactor and may rely on an external redox partner, rather than cofactor exchange, for multiple turnover. Incidentally, these genes always appear in conjunction with the mftA gene, which encodes the short peptide MftA, and with other genes proposed to convert MftA into the external redox partner mycofactocin. -

Bioinformatic Evidence for a Widely Distributed, Ribosomally Produced
Haft BMC Genomics 2011, 12:21 http://www.biomedcentral.com/1471-2164/12/21 RESEARCHARTICLE Open Access Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners Daniel H Haft Abstract Background: Enzymes in the radical SAM (rSAM) domain family serve in a wide variety of biological processes, including RNA modification, enzyme activation, bacteriocin core peptide maturation, and cofactor biosynthesis. Evolutionary pressures and relationships to other cellular constituents impose recognizable grammars on each class of rSAM-containing system, shaping patterns in results obtained through various comparative genomics analyses. Results: An uncharacterized gene cluster found in many Actinobacteria and sporadically in Firmicutes, Chloroflexi, Deltaproteobacteria, and one Archaeal plasmid contains a PqqE-like rSAM protein family that includes Rv0693 from Mycobacterium tuberculosis. Members occur clustered with a strikingly well-conserved small polypeptide we designate “mycofactocin,” similar in size to bacteriocins and PqqA, precursor of pyrroloquinoline quinone (PQQ). Partial Phylogenetic Profiling (PPP) based on the distribution of these markers identifies the mycofactocin cluster, but also a second tier of high-scoring proteins. This tier, strikingly, is filled with up to thirty-one members per genome from three variant subfamilies that occur, one each, in three unrelated classes of nicotinoproteins. The pattern suggests these variant enzymes require not only NAD(P), but also the novel gene cluster. Further study was conducted using SIMBAL, a PPP-like tool, to search these nicotinoproteins for subsequences best correlated across multiple genomes to the presence of mycofactocin. For both the short chain dehydrogenase/reductase (SDR) and iron-containing dehydrogenase families, aligning SIMBAL’s top-scoring sequences to homologous solved crystal structures shows signals centered over NAD(P)-binding sites rather than over substrate-binding or active site residues. -

Functional Characterization of Slscadh1, a Fruit-Ripening Associated Short-Chain Alcohol Dehydrogenase of Tomato
Functional characterization of SlscADH1, a fruit-ripening associated short-chain alcohol dehydrogenase of tomato Hanane Moummou, Libert Brice Tonfack, Christian Chervin, Mohamed Benichou, Emmanuel Youmbi, Christian Ginies, Alain Latché, Jean-Claude Pech, Benoît van der Rest To cite this version: Hanane Moummou, Libert Brice Tonfack, Christian Chervin, Mohamed Benichou, Emmanuel Youmbi, et al.. Functional characterization of SlscADH1, a fruit-ripening associated short-chain alcohol dehy- drogenase of tomato. Journal of Plant Physiology, Elsevier, 2012, vol. 169 (15), pp. 1435-1444. 10.1016/j.jplph.2012.06.007. hal-00780274 HAL Id: hal-00780274 https://hal.archives-ouvertes.fr/hal-00780274 Submitted on 23 Jan 2013 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Open Archive Toulouse Archive Ouverte ( OATAO ) OATAO is an open access repository that collects the work of Toulouse researchers and makes it freely available over the web where possible. This is an author-deposited version published in: http://oatao.univ-toulouse.fr/ Eprints ID : 7966 To link to this article : DOI: 10.1016/j.jplph.2012.06.007 URL: http://dx.doi.org/10.1016/j.jplph.2012.06.007 To cite this version : Moummou, Hanane and Tonfack, Libert Brice and Chervin, Christian and Benichou, Mohamed and Youmbi, Emmanuel and Ginies, Christian and Latché, Alain and Pech, Jean-Claude and Van der Rest, Benoît Functional characterization of SlscADH1, a fruit-ripening associated short-chain alcohol dehydrogenase of tomato.