A Glandular Trichome-Specific Monoterpene Alcohol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Redalyc.Microorganisms Screening for Limonene Oxidation
Ciência e Tecnologia de Alimentos ISSN: 0101-2061 [email protected] Sociedade Brasileira de Ciência e Tecnologia de Alimentos Brasil LERIN, Lindomar; TONIAZZO, Geciane; de OLIVEIRA, Débora; ROTTAVA, Leda; DARIVA, Cláudio; CANSIAN, Rogério Luis; TREICHEL, Helen; PADILHA, Francine; Ceva ANTUNES, Octávio Augusto Microorganisms screening for limonene oxidation Ciência e Tecnologia de Alimentos, vol. 30, núm. 2, abril-junio, 2010, pp. 399-405 Sociedade Brasileira de Ciência e Tecnologia de Alimentos Campinas, Brasil Available in: http://www.redalyc.org/articulo.oa?id=395940100017 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Ciência e Tecnologia de Alimentos ISSN 0101-2061 Microorganisms screening for limonene oxidation Seleção de microrganismos para oxidação de limoneno Original Lindomar LERIN1, Geciane TONIAZZO2, Débora de OLIVEIRA2*, Leda ROTTAVA1, Cláudio DARIVA3, Rogério Luis CANSIAN2, Helen TREICHEL2, Francine PADILHA3, Octávio Augusto Ceva ANTUNES1 Abstract Limonene is a monoterpene obtained in large amounts from essential oils and is used as a raw material for the synthesis of flavors and fine chemicals. Several pathways or routes for the microbial degradation of limonene making use of the cytochrome P450-dependent monooxygenases have been described. In this study, we present a fermentative screening of microorganisms in order to verify their ability to perform the desirable conversion. In parallel, the PCR technique was used to select the microorganisms that contain the limC gene, which is responsible for the conversion of carveol to carvone. -

Enzyme DHRS7
Toward the identification of a function of the “orphan” enzyme DHRS7 Inauguraldissertation zur Erlangung der Würde eines Doktors der Philosophie vorgelegt der Philosophisch-Naturwissenschaftlichen Fakultät der Universität Basel von Selene Araya, aus Lugano, Tessin Basel, 2018 Originaldokument gespeichert auf dem Dokumentenserver der Universität Basel edoc.unibas.ch Genehmigt von der Philosophisch-Naturwissenschaftlichen Fakultät auf Antrag von Prof. Dr. Alex Odermatt (Fakultätsverantwortlicher) und Prof. Dr. Michael Arand (Korreferent) Basel, den 26.6.2018 ________________________ Dekan Prof. Dr. Martin Spiess I. List of Abbreviations 3α/βAdiol 3α/β-Androstanediol (5α-Androstane-3α/β,17β-diol) 3α/βHSD 3α/β-hydroxysteroid dehydrogenase 17β-HSD 17β-Hydroxysteroid Dehydrogenase 17αOHProg 17α-Hydroxyprogesterone 20α/βOHProg 20α/β-Hydroxyprogesterone 17α,20α/βdiOHProg 20α/βdihydroxyprogesterone ADT Androgen deprivation therapy ANOVA Analysis of variance AR Androgen Receptor AKR Aldo-Keto Reductase ATCC American Type Culture Collection CAM Cell Adhesion Molecule CYP Cytochrome P450 CBR1 Carbonyl reductase 1 CRPC Castration resistant prostate cancer Ct-value Cycle threshold-value DHRS7 (B/C) Dehydrogenase/Reductase Short Chain Dehydrogenase Family Member 7 (B/C) DHEA Dehydroepiandrosterone DHP Dehydroprogesterone DHT 5α-Dihydrotestosterone DMEM Dulbecco's Modified Eagle's Medium DMSO Dimethyl Sulfoxide DTT Dithiothreitol E1 Estrone E2 Estradiol ECM Extracellular Membrane EDTA Ethylenediaminetetraacetic acid EMT Epithelial-mesenchymal transition ER Endoplasmic Reticulum ERα/β Estrogen Receptor α/β FBS Fetal Bovine Serum 3 FDR False discovery rate FGF Fibroblast growth factor HEPES 4-(2-Hydroxyethyl)-1-Piperazineethanesulfonic Acid HMDB Human Metabolome Database HPLC High Performance Liquid Chromatography HSD Hydroxysteroid Dehydrogenase IC50 Half-Maximal Inhibitory Concentration LNCaP Lymph node carcinoma of the prostate mRNA Messenger Ribonucleic Acid n.d. -

Biosynthesis of New Alpha-Bisabolol Derivatives Through a Synthetic Biology Approach Arthur Sarrade-Loucheur
Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach Arthur Sarrade-Loucheur To cite this version: Arthur Sarrade-Loucheur. Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach. Biochemistry, Molecular Biology. INSA de Toulouse, 2020. English. NNT : 2020ISAT0003. tel-02976811 HAL Id: tel-02976811 https://tel.archives-ouvertes.fr/tel-02976811 Submitted on 23 Oct 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE En vue de l’obtention du DOCTORAT DE L’UNIVERSITÉ DE TOULOUSE Délivré par l'Institut National des Sciences Appliquées de Toulouse Présentée et soutenue par Arthur SARRADE-LOUCHEUR Le 30 juin 2020 Biosynthèse de nouveaux dérivés de l'α-bisabolol par une approche de biologie synthèse Ecole doctorale : SEVAB - Sciences Ecologiques, Vétérinaires, Agronomiques et Bioingenieries Spécialité : Ingénieries microbienne et enzymatique Unité de recherche : TBI - Toulouse Biotechnology Institute, Bio & Chemical Engineering Thèse dirigée par Gilles TRUAN et Magali REMAUD-SIMEON Jury -
Tropane and Granatane Alkaloid Biosynthesis: a Systematic Analysis
Office of Biotechnology Publications Office of Biotechnology 11-11-2016 Tropane and Granatane Alkaloid Biosynthesis: A Systematic Analysis Neill Kim Texas Tech University Olga Estrada Texas Tech University Benjamin Chavez Texas Tech University Charles Stewart Jr. Iowa State University, [email protected] John C. D’Auria Texas Tech University Follow this and additional works at: https://lib.dr.iastate.edu/biotech_pubs Part of the Biochemical and Biomolecular Engineering Commons, and the Biotechnology Commons Recommended Citation Kim, Neill; Estrada, Olga; Chavez, Benjamin; Stewart, Charles Jr.; and D’Auria, John C., "Tropane and Granatane Alkaloid Biosynthesis: A Systematic Analysis" (2016). Office of Biotechnology Publications. 11. https://lib.dr.iastate.edu/biotech_pubs/11 This Article is brought to you for free and open access by the Office of Biotechnology at Iowa State University Digital Repository. It has been accepted for inclusion in Office of Biotechnology Publicationsy b an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Tropane and Granatane Alkaloid Biosynthesis: A Systematic Analysis Abstract The tropane and granatane alkaloids belong to the larger pyrroline and piperidine classes of plant alkaloids, respectively. Their core structures share common moieties and their scattered distribution among angiosperms suggest that their biosynthesis may share common ancestry in some orders, while they may be independently derived in others. Tropane and granatane alkaloid diversity arises from the myriad modifications occurring ot their core ring structures. Throughout much of human history, humans have cultivated tropane- and granatane-producing plants for their medicinal properties. This manuscript will discuss the diversity of their biological and ecological roles as well as what is known about the structural genes and enzymes responsible for their biosynthesis. -

Effects of a Bacterial ACC Deaminase on Plant Growth
Effects of a bacterial ACC deaminase on plant growth-promotion by Jennifer Claire Czarny A thesis presented to the University of Waterloo in fulfilment of the thesis requirement for the degree of Doctor of Philosophy in Biology Waterloo Ontario, Canada, 2008 c Jennifer Claire Czarny 2008 Author's declaration I hereby declare that I am the sole author of this thesis. This is a true copy of the thesis, including any required final revisions, as accepted by my examiners. I understand that my thesis may be made electronically available to the public. ii Abstract Plants often live in association with growth-promoting bacteria, which provide them with several benefits. One such benefit is the lowering of plant ethylene levels through the action of the bacterial enzyme 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase that cleaves the immediate biosynthetic precursor of ethylene, ACC. The plant hormone ethylene is responsible for many aspects of plant growth and development but under stressful conditions ethylene exacerbates stress symptoms. The ACC deaminase-containing bacterium Pseudomonas putida UW4, isolated from the rhizosphere of reeds, is a potent plant growth- promoting strain and as such was used, along with an ACC deaminase minus mutant of this strain, to study the role of ACC deaminase in plant growth-promotion. Also, transgenic plants expressing a bacterial ACC deaminase gene were used to study the role of this enzyme in plant growth and stress tolerance in the presence and absence of nickel. Transcriptional changes occurring within plant tissues were investigated with the use of an Arabidopsis oligonucleotide microarray. The results showed that transcription of genes involved in hormone regulation, secondary metabolism and the stress response changed in all treatments. -

Metabolic Enzyme/Protease
Inhibitors, Agonists, Screening Libraries www.MedChemExpress.com Metabolic Enzyme/Protease Metabolic pathways are enzyme-mediated biochemical reactions that lead to biosynthesis (anabolism) or breakdown (catabolism) of natural product small molecules within a cell or tissue. In each pathway, enzymes catalyze the conversion of substrates into structurally similar products. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation. Metabolism maintains the living state of the cells and the organism. Proteases are used throughout an organism for various metabolic processes. Proteases control a great variety of physiological processes that are critical for life, including the immune response, cell cycle, cell death, wound healing, food digestion, and protein and organelle recycling. On the basis of the type of the key amino acid in the active site of the protease and the mechanism of peptide bond cleavage, proteases can be classified into six groups: cysteine, serine, threonine, glutamic acid, aspartate proteases, as well as matrix metalloproteases. Proteases can not only activate proteins such as cytokines, or inactivate them such as numerous repair proteins during apoptosis, but also expose cryptic sites, such as occurs with β-secretase during amyloid precursor protein processing, shed various transmembrane proteins such as occurs with metalloproteases and cysteine proteases, or convert receptor agonists into antagonists and vice versa such as chemokine conversions carried out by metalloproteases, dipeptidyl peptidase IV and some cathepsins. In addition to the catalytic domains, a great number of proteases contain numerous additional domains or modules that substantially increase the complexity of their functions. -

Supplementary. Table S1 a Total of Degs Detected in This Study (Gm) No
Supplementary. Table S1 A total of DEGs detected in this study (Gm) No. genename significance in annotation 1 At1g01020 D2 ARV1__expressed protein, similar to hypothetical protein DDB0188786 [Dictyostelium discoideum] (GB:EAL62332.1); contains InterPro domain Arv1-like protein (InterPro:IPR007290) 2 At1g01100 D2 60S acidic ribosomal protein P1 (RPP1A), similar to 60S ACIDIC RIBOSOMAL PROTEIN P1 GB:O23095 from (Arabidopsis thaliana) 3 At1g01120 D2, Dm KCS1__fatty acid elongase 3-ketoacyl-CoA synthase 1 (KCS1), nearly identical to GB:AAC99312 GI:4091810 from (Arabidopsis thaliana) 4 At1g01160 D1, D2, Dm GIF2__SSXT protein-related / transcription co-activator-related, similar to SYT/SSX4 fusion protein (GI:11127695) (Homo sapiens); supporting cDNA gi:21539891:gb:AY102640.1:; contains Pfam profile PF05030: SSXT protein (N-terminal region) 5 At1g01170 D2 ozone-responsive stress-related protein, putative, similar to stress-related ozone-induced protein AtOZI1 (GI:790583) (Arabidopsis thaliana); contains 1 predicted transmembrane domain; 6 At1g01240 D1, D2, Dm expressed protein 7 At1g01300 D2, Dm aspartyl protease family protein, contains Pfam domain, PF00026: eukaryotic aspartyl protease 8 At1g01320 D2 tetratricopeptide repeat (TPR)-containing protein, low similarity to SP:P46825 Kinesin light chain (KLC) {Loligo pealeii}; contains Pfam profile PF00515: TPR Domain 9 At1g01430 D2, Dm expressed protein, similar to hypothetical protein GB:CAB80917 GI:7267605 from (Arabidopsis thaliana) 10 At1g01470 D1, D2, Dm LEA14_LSR3__late embryogenesis abundant -

Investigating Labdane-Related Diterpenoids Biosynthetic Network In
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2012 Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase Yisheng Wu Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry Commons Recommended Citation Wu, Yisheng, "Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase" (2012). Graduate Theses and Dissertations. 12680. https://lib.dr.iastate.edu/etd/12680 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Investigating labdane-related diterpenoids biosynthetic network in rice: Characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase by Yisheng Wu A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Reuben Peters, Major Professor Gustavo MacIntosh Thomas Bobik Bing Yang Mark Hargrove Iowa State University Ames, Iowa 2012 Copyright © Yisheng Wu , 2012. All -

Biocatalysis Using Plant and Metagenomic Enzymes for Organic Synthesis
University College London UCL Biocatalysis Using Plant and Metagenomic Enzymes for Organic Synthesis Sophie Alice Newgas Submitted in partial fulfilment of the requirements for the degree of Doctor of Philosophy (PhD) 2018 [1] [2] Declaration I, Sophie Alice Newgas, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Signed: Dated: [3] Abstract Biocatalysts provide an excellent alternative to traditional organic chemistry strategies, with advantages such as mild reaction conditions and high enantio- and stereoselectivities. The use of metagenomics has enabled new enzymes to be sourced with high sequence diversity. At UCL a metagenomics strategy has been developed for enzyme discovery, in which the library generated is annotated and searched for desired enzyme sequences. In this PhD, a metagenomic approach was used to retrieve 37 short chain reductase/dehydrogenases (SDRs) from an oral environment metagenome. Eight enzymes displayed activity towards cyclohexanone and their substrate selectivities were investigated. Four of the SDRs displayed activity to the Wieland-Miescher ketone (WMK), a motif found in several pharmaceutically relevant compounds. SDR- 17 displayed high conversions and stereoselectivities and was co-expressed with the co-factor recycling enzyme glucose-6-phosphate dehydrogenase. This system was then successfully used to reduce (R)-WMK on a preparative scale reaction in 89% isolated yield and >99% e.e.. In further studies using reductases, the substrate specificities of two ketoreductases known as tropinone reductase I and II (TRI and TRII respectively) from the plant D. stramonium and MecgoR from E. -

Table 3. PDB Representation of Gene Families A. H. Sapiens
Table 3. PDB representation of gene families A. H. -

Pea SAD Short-Chain Dehydrogenase/Reductase Author to Whom All Correspondence Should Be Addressed
Plant Physiology Preview. Published on February 22, 2011, as DOI:10.1104/pp.111.173336 1 Running head: Pea SAD Short-Chain Dehydrogenase/Reductase Author to whom all correspondence should be addressed: Åke Strid Akademin för naturvetenskap och teknik och Centrum för livsvetenskap Örebro universitet SE-70182 Örebro Sweden Telephone: +46-19-303603 E-mail: [email protected] Most appropriate journal research area: Cell Biology 1 Copyright 2011 by the American Society of Plant Biologists 2 The Pisum sativum SAD Short-Chain Dehydrogenase/Reductase: Quinone Reduction, Tissue 1 Distribution, and Heterologous Expression Nikolai Scherbak2, Anneli Ala-Häivälä2, Mikael Brosché2, Nathalie Böwer, Hilja Strid, John R. Gittins3, Elin Grahn, Leif A. Eriksson4, and Åke Strid* Akademin för naturvetenskap och teknik och Centrum för livsvetenskap (N.S., A.A.-H., N.B., E.G., L.A.E., Å.S) and Hälsoakademin och Centrum för livsvetenskap (H.S.), Örebro universitet, S-70182 Örebro, Sweden; Biokemi och biofysik (M.B., J.R.G.), Institutionen för Kemi, Göteborgs universitet, P.O. Box 462, S-405 30 Göteborg, Sweden; Division of Plant Biology (M.B.), Department of Biosciences, University of Helsinki, P.O. Box 65, FIN-00014 Helsinki, Finland, and Institute of Technology, University of Tartu, Nooruse 1, Tartu 50411, Estonia. 2 3 Footnotes: 1This work was supported by Helge Ax:son Johnson’s foundation (N.S.), the Carl Trygger Foundation (Å.S.) and the Faculty for Science and Technology at Örebro University (Å.S.). The Strategic Network for Swedish Plant Biotechnology (SSF) funded J.R.G.’s stay in Sweden with a grant to Å.S. -
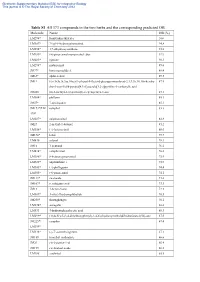
Table S1 All 373 Compounds in the Two Herbs and the Corresponding
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Table S1 All 373 compounds in the two herbs and the corresponding predicted OB Molecule Name OB (%) LM294* forsythidmethylester 100 LM387* 7'-epi-8-hydroxypinoresinol 94.8 LM394* 1,7-dihydroxyxanthone 93.8 LM289* (+)-pinoresinol monomethyl ether 92.9 LM285* egenine 90.3 LM298* matairesinol 89.6 JM77* loniceracetalide A 89.4 JM63* alpha-cedrol 89.3 JM1* (-)-(3r,8s,9r,9as,10as)-9-ethenyl-8-(beta-d-glucopyranosyloxy)-2,3,9,9a,10,10a-hexahy 87.5 dro-5-oxo-5h,8h-pyrano[4,3-d]oxazolo[3,2-a]pyridine-3-carboxylic acid JM200 (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one 87.1 LM304* phillyrin 85.1 JM57* 7-epi-loganin 85.1 JM173*/LM camphol 83.3 398* LM287* epipinoresinol 82.8 JM27 2-methyl-1-butanol 81.2 LM356* (+)-lariciresinol 80.0 JM170* ledol 79.7 LM430 safynol 78.3 JM16 1-pentanol 76.2 LM414* campherenol 76.0 LM386* 8-hydroxypinoresinol 75.9 LM428* onjixanthone i 75.9 LM345* (+)-phillygenin 74.4 LM305* (+)-pinoresinol 74.1 JM113* sweroside 73.6 JM107* secologanic acid 73.3 JM13 1-hexen-3-one 72.3 LM309* 3-ethyl-7hydroxyphthalide 70.5 JM250* shuangkangsu 70.1 LM274* astragalin 68.6 LM331 4-hydroxyphenylacetic acid 68.3 LM299* (3r,4s,5r)-5-(3,4-dimethoxyphenyl)-3,4-bis(hydroxymethyl)dihydrofuran-2(3h)-one 67.5 JM225*/ camphor 67.4 LM399* LM338* (-)-7’-o-methylegenine 67.1 JM189 trimethyl isothiazole 66.6 JM29 cis-2-penten-1-ol 66.4 JM199 cis-linalool oxide 66.2 LM288 erythritol 65.8 Electronic Supplementary Material (ESI) for