Using a Bioluminescent Bacterial Bioreporter to Assess Iron Bioavailability in the Oceans
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporter Genes a Practical Guide M E T H O D S I N M O L E C U L a R B I O L O G Y™
Reporter Genes A Practical Guide M E T H O D S I N M O L E C U L A R B I O L O G Y™ John M. Walker, SERIES EDITOR 436. Avian Influenza Virus, edited by 413. Protein Structure Prediction, Second Edition, Erica Spackman, 2008 edited by Mohammed Zaki and Chris Bystroff, 435. Chromosomal Mutagenesis, edited by 2008 Greg Davis and Kevin J. Kayser, 2008 412. Neutrophil Methods and Protocols, edited by 434. Gene Therapy Protocols: Vol. 2: Design and Mark T. Quinn, Frank R. DeLeo, Characterization of Gene Transfer Vectors, and Gary M. Bokoch, 2007 edited by Joseph M. LeDoux, 2008 411. Reporter Genes: A Practical Guide, edited by 433. Gene Therapy Protocols: Vol. 1: Production Don Anson, 2007 and In Vivo Applications of Gene Transfer 410. Environmental Genomics, edited by Vectors, edited by Joseph M. LeDoux, 2008 Cristofre C. Martin, 2007 432. Organelle Proteomics, edited by 409. Immunoinformatics: Predicting Immunogenicity Delphine Pflieger and Jean Rossier, 2008 In Silico, edited by Darren R. Flower, 2007 431. Bacterial Pathogenesis: Methods and 408. Gene Function Analysis, edited by Protocols, edited by Frank DeLeo and Michael Ochs, 2007 Michael Otto, 2008 407. Stem Cell Assays, edited by Vemuri C. Mohan, 430. Hematopoietic Stem Cell Protocols, 2007 edited by Kevin D. Bunting, 2008 406. Plant Bioinformatics: Methods and Protocols, 429. Molecular Beacons: Signalling Nucleic Acid edited by David Edwards, 2007 Probes, Methods and Protocols, edited by 405. Telomerase Inhibition: Strategies and Andreas Marx and Oliver Seitz, 2008 Protocols, edited by Lucy Andrews and 428. Clinical Proteomics: Methods and Protocols, Trygve O. -

FDA: "Glowing" Seafood?
FDA: "Glowing" Seafood? http://web.archive.org/web/20080225162926/http://vm.cfsan.fda.gov/~ea... U.S. Food and Drug Administration Seafood Products Research Center July 1998 "GLOWING" SEAFOOD? by Patricia N. Sado* Introduction Seafood that produces a bright, blue-green light in the dark could be a meal from outer space or haute cuisine in a science fiction novel. The U. S. Food and Drug Administration (FDA) has received many consumer complaints about various seafood products "glowing" in the dark. Some of these consumers called their local health departments, poison control centers, and their U.S. Senator because they thought they had been poisoned by radiation. These consumers said they had trouble convincing people that their seafood was emitting light. One consumer took his imitation crabmeat to a local television station. Unfortunately his seafood had dried out and did not glow for the television reporters. Several consumers said that it took them many weeks before they found phone numbers for various government agencies to make inquiries. Several consumers thought their "glowing" seafood was due to phosphorescing phytoplankton, or even fluorescence. The consumers' seafood products "glowing" in the dark were not due to radiation or to fluorescence, which requires an ultraviolet light to trigger the reaction. These seafood products exhibited luminescence due to the presence of certain bacteria that are capable of emitting light. Luminescence by bacteria is due to a chemical reaction catalyzed by luciferase, a protein similar to that found in fireflies. The reaction involves oxidation of a reduced flavin mononucleotide and a long chain aliphatic aldehyde by molecular oxygen to produce oxidized flavin plus fatty acid and light (5, 12). -

Developments in Aquatic Microbiology
INTERNATL MICROBIOL (2000) 3: 203–211 203 © Springer-Verlag Ibérica 2000 REVIEW ARTICLE Samuel P. Meyers Developments in aquatic Department of Oceanography and Coastal Sciences, Louisiana State University, microbiology Baton Rouge, LA, USA Received 30 August 2000 Accepted 29 September 2000 Summary Major discoveries in marine microbiology over the past 4-5 decades have resulted in the recognition of bacteria as a major biomass component of marine food webs. Such discoveries include chemosynthetic activities in deep-ocean ecosystems, survival processes in oligotrophic waters, and the role of microorganisms in food webs coupled with symbiotic relationships and energy flow. Many discoveries can be attributed to innovative methodologies, including radioisotopes, immunofluores- cent-epifluorescent analysis, and flow cytometry. The latter has shown the key role of marine viruses in marine system energetics. Studies of the components of the “microbial loop” have shown the significance of various phagotrophic processes involved in grazing by microinvertebrates. Microbial activities and dissolved organic carbon are closely coupled with the dynamics of fluctuating water masses. New biotechnological approaches and the use of molecular biology techniques still provide new and relevant information on the role of microorganisms in oceanic and estuarine environments. International interdisciplinary studies have explored ecological aspects of marine microorganisms and their significance in biocomplexity. Studies on the Correspondence to: origins of both life and ecosystems now focus on microbiological processes in the Louisiana State University Station. marine environment. This paper describes earlier and recent discoveries in marine Post Office Box 19090-A. Baton Rouge, LA 70893. USA (aquatic) microbiology and the trends for future work, emphasizing improvements Tel.: +1-225-3885180 in methodology as major catalysts for the progress of this broadly-based field. -

Diversity and Evolution of Bacterial Bioluminescence Genes in the Global Ocean Thomas Vannier, Pascal Hingamp, Floriane Turrel, Lisa Tanet, Magali Lescot, Y
Diversity and evolution of bacterial bioluminescence genes in the global ocean Thomas Vannier, Pascal Hingamp, Floriane Turrel, Lisa Tanet, Magali Lescot, Y. Timsit To cite this version: Thomas Vannier, Pascal Hingamp, Floriane Turrel, Lisa Tanet, Magali Lescot, et al.. Diversity and evolution of bacterial bioluminescence genes in the global ocean. NAR Genomics and Bioinformatics, Oxford University Press, 2020, 2 (2), 10.1093/nargab/lqaa018. hal-02514159 HAL Id: hal-02514159 https://hal.archives-ouvertes.fr/hal-02514159 Submitted on 21 Mar 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Published online 14 March 2020 NAR Genomics and Bioinformatics, 2020, Vol. 2, No. 2 1 doi: 10.1093/nargab/lqaa018 Diversity and evolution of bacterial bioluminescence genes in the global ocean Thomas Vannier 1,2,*, Pascal Hingamp1,2, Floriane Turrel1, Lisa Tanet1, Magali Lescot 1,2,* and Youri Timsit 1,2,* 1Aix Marseille Univ, Universite´ de Toulon, CNRS, IRD, MIO UM110, 13288 Marseille, France and 2Research / Federation for the study of Global Ocean Systems Ecology and Evolution, FR2022 Tara GOSEE, 3 rue Michel-Ange, Downloaded from https://academic.oup.com/nargab/article-abstract/2/2/lqaa018/5805306 by guest on 21 March 2020 75016 Paris, France Received October 21, 2019; Revised February 14, 2020; Editorial Decision March 02, 2020; Accepted March 06, 2020 ABSTRACT ganisms and is particularly widespread in marine species (7–9). -

Construction of a Copper Bioreporter Screening, Characterization and Genetic Improvement of Copper-Sensitive Bacteria
Construction of a Copper Bioreporter Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath This thesis comprises 30 ECTS credits and is a compulsory part in the Master of Science With a Major in Industrial Biotechnology, 120 ECTS credits No. 9/2009 Construction of a Copper Bioreporter-Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath, [email protected] Master thesis Subject Category: Technology University College of Borås School of Engineering SE-501 90 BORÅS Telephone +46 033 435 4640 Examiner: Dr. Elisabeth Feuk-lagerstedt Supervisor, name: Dr. Saman Hosseinkhani Supervisor, address: Department of Biochemistry, Faculty of Biological Sciences, Tarbiat Modares University Tehran, Iran Date: 2009-12-14 Keywords: Copper Bioreporter, Luciferase assay, COP operon, pGL3, E. Coli BL-21 ii Dedicated to my parents to whom I owe and feel the deepest gratitude iii Acknowledgment I would like to express my best appreciation to my supervisor Dr. S. Hosseinkhani for technical supports, and my examiner Dr. E. Feuk-lagerstedt for her kind attention, also I want to thank Mr. A. Emamzadeh, Miss. M. Nazari, and other students of Biochemistry laboratory of Tarbiat Modares University for their kind corporations. iv Abstract In the nature, lots of organism applies different kinds of lights such as flourescence or luminescence for some purposes such as defense or hunting. Firefly luciferase and Bacterial luciferase are the most famous ones which have been used to design Biosensors or Bioreporters in recent decades. Their applications are so extensive from detecting pollutions in the environment to medical and treatment usages. -

New Naphthalene Whole-Cell Bioreporter for Measuring and Assessing Naphthalene in Polycyclic Aromatic Hydrocarbons Contaminated Site
Accepted Manuscript New naphthalene whole-cell bioreporter for measuring and assessing naphthalene in polycyclic aromatic hydrocarbons contaminated site Yujiao Sun, Xiaohui Zhao, Dayi Zhang, Aizhong Ding, Cheng Chen, Wei E. Huang, Huichun Zhang PII: S0045-6535(17)31248-1 DOI: 10.1016/j.chemosphere.2017.08.027 Reference: CHEM 19725 To appear in: ECSN Received Date: 17 April 2017 Revised Date: 22 July 2017 Accepted Date: 7 August 2017 Please cite this article as: Sun, Y., Zhao, X., Zhang, D., Ding, A., Chen, C., Huang, W.E., Zhang, H., New naphthalene whole-cell bioreporter for measuring and assessing naphthalene in polycyclic aromatic hydrocarbons contaminated site, Chemosphere (2017), doi: 10.1016/j.chemosphere.2017.08.027. This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. ACCEPTED MANUSCRIPT 1 New naphthalene whole-cell bioreporter for measuring and assessing 2 naphthalene in polycyclic aromatic hydrocarbons contaminated site 3 Yujiao Sun a, Xiaohui Zhao a,b* , Dayi Zhang c, Aizhong Ding a, Cheng Chen a, Wei E. 4 Huang d, Huichun Zhang a. 5 a College of Water Sciences, Beijing Normal University, Beijing, 100875, PR China 6 b Department of Water Environment, China Institute of Water Resources and 7 Hydropower Research, Beijing, 100038, China 8 c Lancaster Environment Centre, Lancaster University, Lancaster, LA1 4YQ, UK 9 d Kroto Research Institute, University of Sheffield, Sheffield, S3 7HQ, United 10 Kingdom 11 12 Corresponding author 13 Dr Xiaohui Zhao 14 a College of Water Sciences, Beijing Normal UniversiMANUSCRIPTty, Beijing 100875, P. -

Short Communication Isolation of Bioluminescent Bacteria From
Indian Journal of Geo Marine Sciences Vol. 49 (03), March 2020, pp. 471-476 Short Communication Isolation of bioluminescent bacteria from widely distributed light emitting organisms are marine organisms luminescent bacteria. Such bioluminescent bacteria are either found in planktonic form in the ocean6-12 or Paritosh Parmar, Arpit Shukla, Meenu Saraf & Baldev Patel* in symbiotic relationship with the marine host for instance certain fishes and squids2,8,13-17. Department of Microbiology & Biotechnology, University School of Sciences, Gujarat University, Ahmedabad-380 009, India The reaction which is responsible for emission of *[E-mail: [email protected]] light involves the enzyme luciferase that oxidizes long chain aliphatic aldehyde (RCHO), reduced flavin Received 31 July 2018; 05 October 2018 mononucleotide (FMNH2), and oxygen (O2) to a long chain aliphatic acid (RCOOH), oxidized flavin Bioluminescence is an emission of cold light by enzyme driven reaction within certain living organisms. The most mononucleotide (FMN), and water (H2O). In this abundant and widely distributed light emitting organisms are process, the excess energy is liberated and emitted as luminescent bacteria. Such organisms are either found as free- luminescent blue-green light of 490 nm (Fig. 1)4. All living in the ocean or in symbiotic relationship with the marine luminescent bacteria are distributed among three host. To employ bioluminescence in environmental monitoring, 4,18 isolation of bioluminescent bacteria from the two different marine genera Vibrio, Photobacterium, Xenorhabdus . samples (sea water sample and various organs of squid and fish) However, recently one more genera Kurthia has also were collected from different sites of Veraval seashore and fish been reported to be luminescent19. -

Reporter Gene Assays
TCA-017 Reporter Gene Assays Abstract foreign to mammalian cells, and are biologically active without post-translational modification. Luminescent reporter gene assays provide a simple, fast, non-isotopic, and highly sensitive alternative to The high quantum efficiency of luciferase enables the popular radioactive chloramphenicol detection of less than 0.01 attomoles of luciferase, acetyltransferase (CAT) assay. This paper describes making it possible to scale down the assay to the a reporter gene assay in which cells were transfected 96-well microplate format.1,2 Although the luciferin/ with the firefly luciferase gene, which was quanti- luciferase reaction is often thought of as a flash fied using an enhanced flash luciferin substrate chemistry, recent advances have extended the pho- and the TopCount Microplate Scintillation and ton kinetics by the use of coenzyme A as a substrate Luminescence Counter. Co-transfection with the in the oxidation of luciferin.3 The resulting enhanced ß-galactosidase gene may be included to control for flash eliminates the need for elaborate reagent inter-assay transfection efficiency. Both of these injection devices. luminescent reporters can be quantified without reagent injection devices. The signal decays only slightly during the time required to read a microplate, Co-transfection with a second reporter gene is often and can be easily corrected for the decay by tandem performed to normalize transfection efficiency processing on TopCount. between assays. The ß-galactosidase gene is ideal for this application. Chemiluminescent substrates for ß- galactosidase are sensitive enough to allow detec- Introduction tion at low transfection efficiency, are available in compatible plasmid vectors, and are easily adaptable The use of reporter genes is a fundamental tool of 4 molecular biologists. -
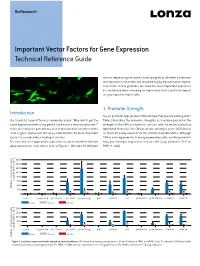
Important Vector Factors for Gene Expression Technical Reference Guide
BioResearch Important Vector Factors for Gene Expression Technical Reference Guide vectors expressing the same luciferase gene in different backbones and expression cassettes and obtained highly discriminative expres- sion levels. In this guideline, we show the most important aspects to be considered when choosing an expression vector and their impact on your experimental results. 1. Promoter Strength Introduction Is your promoter appropriate for the cell type that you are working with? Our Scientific Support Team is commonly asked: “Why don’t I get the Table 1 describes the promoter strengths as a relative percent of the same expression level of my gene if I use various vector backbones?” strength of the CMV promoter for various cells for which Lonza has There are many components to a vector that can have an effect on the Optimized Protocols. The CMV promoter activity is set to 100% based level of gene expression. Below you will find the 10 most important on their CAT assay values from the referenced publications. Although factors to consider when looking at vectors. CMV is a strong promoter in many mammalian cells, another promoter The selection of an appropriate expression vector is crucial for efficient may give stronger expression in your cells (e.g., promoter SV40 in gene expression. Just take a look at Figure 1. We tried 10 different BHK-21 cells). n 1600 1400 1200 1000 800 [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 600 400 Relative luciferase-expressio Relative 200 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT 4 hours 24 hours 48 hours n 2000 1500 1000 500 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 4 hours 24 hours 48 hours Relative luciferase-expressio Relative Figure 1: Luciferase expression levels depend on vector backbones. -

Robust Normalization of Luciferase Reporter Data
Technical Note Robust Normalization of Luciferase Reporter Data Andrea Repele † and Manu * Department of Biology, University of North Dakota, Grand Forks, ND 58202, USA * Correspondence: [email protected] † Current address: JMB—Center for Immunity and Immunotherapies, Seattle Children’s Hospital, Seattle, WA 98105, USA. Received: 17 June 2019; Accepted: 22 July 2019; Published: 25 July 2019 Abstract: Transient Luciferase reporter assays are widely used in the study of gene regulation and intracellular cell signaling. In order to control for sample-to-sample variation in luminescence arising from variability in transfection efficiency and other sources, an internal control reporter is co-transfected with the experimental reporter. The luminescence of the experimental reporter is normalized against the control by taking the ratio of the two. Here we show that this method of normalization, “ratiometric”, performs poorly when the transfection efficiency is low and leads to biased estimates of relative activity. We propose an alternative methodology based on linear regression that is much better suited for the normalization of reporter data, especially when transfection efficiency is low. We compare the ratiometric method against three regression methods on both simulated and empirical data. Our results suggest that robust errors-in-variables (REIV) regression performs the best in normalizing Luciferase reporter data. We have made the R code for Luciferase data normalization using REIV available on GitHub. Keywords: luciferase reporter; transfection efficiency; normalization; gene regulation; promoter; enhancer 1. Introduction Transient reporter assays are an important and widely used tool in the study of gene regulation [1–4], intracellular cell signaling [5–7], and other areas of molecular, cellular, and developmental biology [8–10]. -

Isolation and Identification of Bioluminescent Bacteria in Squid and Water of Malaysia
Int'l Journal of Advances in Agricultural & Environmental Engg. (IJAAEE) Vol. 1, Issue 2 (2014) ISSN 2349-1523 EISSN 2349-1531 Isolation And Identification Of Bioluminescent Bacteria In Squid And Water Of Malaysia Noor Aini Yaser1, Mohd Faiz Foong Abdullah2, Aslizah Mohd Aris3, Iwana Izni Zainudin3 also in marine ecosystem. Abstract— Bioluminescence is an emission of light that produce These bioluminescent bacteria reported belong to the class by chemical reaction of enzymatic activity and biochemical of the Gammaproteobacteria. Among the bioluminescent bacteria living organism. The most abundant and widely distributed light reported, 17 bioluminescent species were currently known [7]. emitting organism is luminous bacteria either found as free-living in The luminous bacteria were also classified into three genera the ocean or gut symbiont in the host. These sets of species are diversely spread in terrestrial environment, freshwater and also in which are Vibrio, Photobacterium and Xenorhabdus marine ecosystem. These species of bacteria emit blue-green light (Photorhabdus) and the species within the genus Vibrio and provoked by the enzyme luciferase and regulated by the lux gene Photobacterium are usually exist in marine environment and operon and emitted at 490 nm. Some of the uses of the Xenorhabdus belong to terrestrial environment. bioluminescence are as genetic reporters and biosensors. General Some of the natural important of the bioluminescence in microbiological techniques have been used to isolate bioluminescent nature is in attracting prey, camouflage, deterring predators bacteria from water samples and squid. The characterization of 6 isolates bioluminescent bacteria was conducted macroscopic and and in aiding in hunting [8]. microscopically and was further analyses at molecular level using Bioluminescence is the phenomenon that consists of light 16sDNA and sequenced for species identification. -

Shedding Light on the Bioluminescence “Paradox” Although Luminescence Provides Host Squids with Obvious Advantages, How Does It Benefit Light-Producing Bacteria?
Shedding Light on the Bioluminescence “Paradox” Although luminescence provides host squids with obvious advantages, how does it benefit light-producing bacteria? Eric V. Stabb he fascinating biochemistry, genetics, have special significance for this bacterium. Bi- and cell density-dependent regula- oluminescence offers many such puzzles, and it tion of bacterial bioluminescence is unlikely a single solution will solve them all. T provoke a challenging question. Nonetheless, it is an exciting moment in bio- What good is it to bioluminescent luminescence research, with recent advances of- bacteria? fering the promise of answering the longstand- There may be no single answer to this seem- ing question, “how can bioluminescence ingly simple question. However, two recent ad- help bacteria?” Although researchers learned vances shed new light on the problem. First, nearly a century ago that luminescence reduces studies of the symbiosis between the biolumines- oxygen and that symbiotic bacteria inhabit the cent bacterium Vibrio fischeri and the Hawaiian light organs of squids, what was unimaginable bobtail squid bring an ecologically relevant until very recently is our ability to analyze the V. niche for a bioluminescent bacterium into focus fischeri genome sequence and to combine this under the discerning lens of controlled labora- knowledge with the ability to genetically manip- tory experimentation. Second, progress in under- ulate these bacteria and observe them under standing the genetics of V. fischeri, including controlled laboratory conditions in the ecologi- genomic sequencing of a squid symbiont, is en- cally relevant environment of a natural squid abling researchers to analyze how luminescence host. integrates into the physiology of this bacterial species and to test specific hypotheses about Biochemistry and Genetics of what advantage light production confers on the V.