Robust Normalization of Luciferase Reporter Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporter Genes a Practical Guide M E T H O D S I N M O L E C U L a R B I O L O G Y™
Reporter Genes A Practical Guide M E T H O D S I N M O L E C U L A R B I O L O G Y™ John M. Walker, SERIES EDITOR 436. Avian Influenza Virus, edited by 413. Protein Structure Prediction, Second Edition, Erica Spackman, 2008 edited by Mohammed Zaki and Chris Bystroff, 435. Chromosomal Mutagenesis, edited by 2008 Greg Davis and Kevin J. Kayser, 2008 412. Neutrophil Methods and Protocols, edited by 434. Gene Therapy Protocols: Vol. 2: Design and Mark T. Quinn, Frank R. DeLeo, Characterization of Gene Transfer Vectors, and Gary M. Bokoch, 2007 edited by Joseph M. LeDoux, 2008 411. Reporter Genes: A Practical Guide, edited by 433. Gene Therapy Protocols: Vol. 1: Production Don Anson, 2007 and In Vivo Applications of Gene Transfer 410. Environmental Genomics, edited by Vectors, edited by Joseph M. LeDoux, 2008 Cristofre C. Martin, 2007 432. Organelle Proteomics, edited by 409. Immunoinformatics: Predicting Immunogenicity Delphine Pflieger and Jean Rossier, 2008 In Silico, edited by Darren R. Flower, 2007 431. Bacterial Pathogenesis: Methods and 408. Gene Function Analysis, edited by Protocols, edited by Frank DeLeo and Michael Ochs, 2007 Michael Otto, 2008 407. Stem Cell Assays, edited by Vemuri C. Mohan, 430. Hematopoietic Stem Cell Protocols, 2007 edited by Kevin D. Bunting, 2008 406. Plant Bioinformatics: Methods and Protocols, 429. Molecular Beacons: Signalling Nucleic Acid edited by David Edwards, 2007 Probes, Methods and Protocols, edited by 405. Telomerase Inhibition: Strategies and Andreas Marx and Oliver Seitz, 2008 Protocols, edited by Lucy Andrews and 428. Clinical Proteomics: Methods and Protocols, Trygve O. -

Casein Kinase 1 Isoforms in Degenerative Disorders
CASEIN KINASE 1 ISOFORMS IN DEGENERATIVE DISORDERS DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Theresa Joseph Kannanayakal, M.Sc., M.S. * * * * * The Ohio State University 2004 Dissertation Committee: Approved by Professor Jeff A. Kuret, Adviser Professor John D. Oberdick Professor Dale D. Vandre Adviser Professor Mike X. Zhu Biophysics Graduate Program ABSTRACT Casein Kinase 1 (CK1) enzyme is one of the largest family of Serine/Threonine protein kinases. CK1 has a wide distribution spanning many eukaryotic families. In cells, its kinase activity has been found in various sub-cellular compartments enabling it to phosphorylate many proteins involved in cellular maintenance and disease pathogenesis. Tau is one such substrate whose hyperphosphorylation results in degeneration of neurons in Alzheimer’s disease (AD). AD is a slow neuroprogessive disorder histopathologically characterized by Granulovacuolar degeneration bodies (GVBs) and intraneuronal accumulation of tau in Neurofibrillary Tangles (NFTs). The level of CK1 isoforms, CK1α, CK1δ and CK1ε has been shown to be elevated in AD. Previous studies of the correlation of CK1δ with lesions had demonstrated its importance in tau hyperphosphorylation. Hence we investigated distribution of CK1α and CK1ε with the lesions to understand if they would play role in tau hyperphosphorylation similar to CK1δ. The kinase results were also compared with lesion correlation studies of peptidyl cis/trans prolyl isomerase (Pin1) and caspase-3. Our results showed that among the enzymes investigated, CK1 isoforms have the greatest extent of colocalization with the lesions. We have also investigated the distribution of CK1α with different stages of NFTs that follow AD progression. -

CKI and CKII Mediate the FREQUENCY-Dependent Phosphorylation of the WHITE COLLAR Complex to Close the Neurospora Circadian Negative Feedback Loop
Downloaded from genesdev.cshlp.org on September 24, 2021 - Published by Cold Spring Harbor Laboratory Press CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop Qun He,1,2 Joonseok Cha,1 Qiyang He,1,3 Heng-Chi Lee,1 Yuhong Yang,1,4 and Yi Liu1,5 1Department of Physiology, The University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA; 2State Key Laboratory for Agro-Biotechnology, College of Biological Sciences, China Agricultural University, Beijing 100094, China The eukaryotic circadian oscillators consist of circadian negative feedback loops. In Neurospora,itwas proposed that the FREQUENCY (FRQ) protein promotes the phosphorylation of the WHITE COLLAR (WC) complex, thus inhibiting its activity. The kinase(s) involved in this process is not known. In this study, we show that the disruption of the interaction between FRQ and CK-1a (a casein kinase I homolog) results in the hypophosphorylation of FRQ, WC-1, and WC-2. In the ck-1aL strain, a knock-in mutant that carries a mutation equivalent to that of the Drosophila dbtL mutation, FRQ, WC-1, and WC-2 are hypophosphorylated. The mutant also exhibits ∼32 h circadian rhythms due to the increase of FRQ stability and the significant delay of FRQ progressive phosphorylation. In addition, the levels of WC-1 and WC-2 are low in the ck-1aL strain, indicating that CK-1a is also important for the circadian positive feedback loops. In spite of its low accumulation in the ck-1aL strain, the hypophosphorylated WCC efficiently binds to the C-box within the frq promoter, presumably because it cannot be inactivated through FRQ-mediated phosphorylation. -

Construction of a Copper Bioreporter Screening, Characterization and Genetic Improvement of Copper-Sensitive Bacteria
Construction of a Copper Bioreporter Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath This thesis comprises 30 ECTS credits and is a compulsory part in the Master of Science With a Major in Industrial Biotechnology, 120 ECTS credits No. 9/2009 Construction of a Copper Bioreporter-Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath, [email protected] Master thesis Subject Category: Technology University College of Borås School of Engineering SE-501 90 BORÅS Telephone +46 033 435 4640 Examiner: Dr. Elisabeth Feuk-lagerstedt Supervisor, name: Dr. Saman Hosseinkhani Supervisor, address: Department of Biochemistry, Faculty of Biological Sciences, Tarbiat Modares University Tehran, Iran Date: 2009-12-14 Keywords: Copper Bioreporter, Luciferase assay, COP operon, pGL3, E. Coli BL-21 ii Dedicated to my parents to whom I owe and feel the deepest gratitude iii Acknowledgment I would like to express my best appreciation to my supervisor Dr. S. Hosseinkhani for technical supports, and my examiner Dr. E. Feuk-lagerstedt for her kind attention, also I want to thank Mr. A. Emamzadeh, Miss. M. Nazari, and other students of Biochemistry laboratory of Tarbiat Modares University for their kind corporations. iv Abstract In the nature, lots of organism applies different kinds of lights such as flourescence or luminescence for some purposes such as defense or hunting. Firefly luciferase and Bacterial luciferase are the most famous ones which have been used to design Biosensors or Bioreporters in recent decades. Their applications are so extensive from detecting pollutions in the environment to medical and treatment usages. -

Reporter Gene Assays
TCA-017 Reporter Gene Assays Abstract foreign to mammalian cells, and are biologically active without post-translational modification. Luminescent reporter gene assays provide a simple, fast, non-isotopic, and highly sensitive alternative to The high quantum efficiency of luciferase enables the popular radioactive chloramphenicol detection of less than 0.01 attomoles of luciferase, acetyltransferase (CAT) assay. This paper describes making it possible to scale down the assay to the a reporter gene assay in which cells were transfected 96-well microplate format.1,2 Although the luciferin/ with the firefly luciferase gene, which was quanti- luciferase reaction is often thought of as a flash fied using an enhanced flash luciferin substrate chemistry, recent advances have extended the pho- and the TopCount Microplate Scintillation and ton kinetics by the use of coenzyme A as a substrate Luminescence Counter. Co-transfection with the in the oxidation of luciferin.3 The resulting enhanced ß-galactosidase gene may be included to control for flash eliminates the need for elaborate reagent inter-assay transfection efficiency. Both of these injection devices. luminescent reporters can be quantified without reagent injection devices. The signal decays only slightly during the time required to read a microplate, Co-transfection with a second reporter gene is often and can be easily corrected for the decay by tandem performed to normalize transfection efficiency processing on TopCount. between assays. The ß-galactosidase gene is ideal for this application. Chemiluminescent substrates for ß- galactosidase are sensitive enough to allow detec- Introduction tion at low transfection efficiency, are available in compatible plasmid vectors, and are easily adaptable The use of reporter genes is a fundamental tool of 4 molecular biologists. -
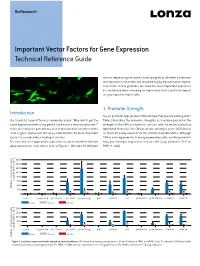
Important Vector Factors for Gene Expression Technical Reference Guide
BioResearch Important Vector Factors for Gene Expression Technical Reference Guide vectors expressing the same luciferase gene in different backbones and expression cassettes and obtained highly discriminative expres- sion levels. In this guideline, we show the most important aspects to be considered when choosing an expression vector and their impact on your experimental results. 1. Promoter Strength Introduction Is your promoter appropriate for the cell type that you are working with? Our Scientific Support Team is commonly asked: “Why don’t I get the Table 1 describes the promoter strengths as a relative percent of the same expression level of my gene if I use various vector backbones?” strength of the CMV promoter for various cells for which Lonza has There are many components to a vector that can have an effect on the Optimized Protocols. The CMV promoter activity is set to 100% based level of gene expression. Below you will find the 10 most important on their CAT assay values from the referenced publications. Although factors to consider when looking at vectors. CMV is a strong promoter in many mammalian cells, another promoter The selection of an appropriate expression vector is crucial for efficient may give stronger expression in your cells (e.g., promoter SV40 in gene expression. Just take a look at Figure 1. We tried 10 different BHK-21 cells). n 1600 1400 1200 1000 800 [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 600 400 Relative luciferase-expressio Relative 200 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT 4 hours 24 hours 48 hours n 2000 1500 1000 500 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 4 hours 24 hours 48 hours Relative luciferase-expressio Relative Figure 1: Luciferase expression levels depend on vector backbones. -

Pressure Accelerates the Circadian Clock of Cyanobacteria
www.nature.com/scientificreports OPEN Pressure accelerates the circadian clock of cyanobacteria Ryo Kitahara 1,2, Katsuaki Oyama2, Takahiro Kawamura2, Keita Mitsuhashi2, Soichiro Kitazawa1, Kazuhiro Yasunaga1, Natsuno Sagara1, Megumi Fujimoto2 & Kazuki Terauchi2,3 Received: 12 April 2019 Although organisms are exposed to various pressure and temperature conditions, information remains Accepted: 7 August 2019 limited on how pressure afects biological rhythms. This study investigated how hydrostatic pressure Published: xx xx xxxx afects the circadian clock (KaiA, KaiB, and KaiC) of cyanobacteria. While the circadian rhythm is inherently robust to temperature change, KaiC phosphorylation cycles that were accelerated from 22 h at 1 bar to 14 h at 200 bars caused the circadian-period length to decline. This decline was caused by the pressure-induced enhancement of KaiC ATPase activity and allosteric efects. Because ATPase activity was elevated in the CI and CII domains of KaiC, while ATP hydrolysis had negative activation volumes (ΔV≠), both domains played key roles in determining the period length of the KaiC phosphorylation cycle. The thermodynamic contraction of the structure of the active site during the transition state might have positioned catalytic residues and lytic water molecules favourably to facilitate ATP hydrolysis. Internal cavities might represent sources of compaction and structural rearrangement in the active site. Overall, the data indicate that pressure diferences could alter the circadian rhythms of diverse organisms with evolved thermotolerance, as long as enzymatic reactions defning period length have a specifc activation volume. Circadian rhythms are endogenous timing systems that induce the circadian clock, resulting in numerous organisms, from cyanobacteria to higher animals, being adapted to the day-night cycle1,2. -

I = Chpt 15. Positive and Negative Transcriptional Control at Lac BMB
BMB 400 Part Four - I = Chpt 15. Positive and Negative Transcriptional Control at lac B M B 400 Part Four: Gene Regulation Section I = Chapter 15 POSITIVE AND NEGATIVE CONTROL SHOWN BY THE lac OPERON OF E. COLI A. Definitions and general comments 1. Operons An operon is a cluster of coordinately regulated genes. It includes structural genes (generally encoding enzymes), regulatory genes (encoding, e.g. activators or repressors) and regulatory sites (such as promoters and operators). 2. Negative versus positive control a. The type of control is defined by the response of the operon when no regulatory protein is present. b. In the case of negative control, the genes in the operon are expressed unless they are switched off by a repressor protein. Thus the operon will be turned on constitutively (the genes will be expressed) when the repressor in inactivated. c. In the case of positive control, the genes are expressed only when an active regulator protein, e.g. an activator, is present. Thus the operon will be turned off when the positive regulatory protein is absent or inactivated. Table 4.1.1. Positive vs. negative control BMB 400 Part Four - I = Chpt 15. Positive and Negative Transcriptional Control at lac 3. Catabolic versus biosynthetic operons a. Catabolic pathways catalyze the breakdown of nutrients (the substrate for the pathway) to generate energy, or more precisely ATP, the energy currency of the cell. In the absence of the substrate, there is no reason for the catabolic enzymes to be present, and the operon encoding them is repressed. In the presence of the substrate, when the enzymes are needed, the operon is induced or de-repressed. -

Analysis of Codon Usage Patterns in Giardia Duodenalis Based on Transcriptome Data from Giardiadb
G C A T T A C G G C A T genes Article Analysis of Codon Usage Patterns in Giardia duodenalis Based on Transcriptome Data from GiardiaDB Xin Li, Xiaocen Wang, Pengtao Gong, Nan Zhang, Xichen Zhang and Jianhua Li * Key Laboratory of Zoonosis Research, Ministry of Education, College of Veterinary Medicine, Jilin University, Changchun 130062, China; [email protected] (X.L.); [email protected] (X.W.); [email protected] (P.G.); [email protected] (N.Z.); [email protected] (X.Z.) * Correspondence: [email protected]; Tel.: +86-431-8783-6172; Fax: +86-431-8798-1351 Abstract: Giardia duodenalis, a flagellated parasitic protozoan, the most common cause of parasite- induced diarrheal diseases worldwide. Codon usage bias (CUB) is an important evolutionary character in most species. However, G. duodenalis CUB remains unclear. Thus, this study analyzes codon usage patterns to assess the restriction factors and obtain useful information in shaping G. duo- denalis CUB. The neutrality analysis result indicates that G. duodenalis has a wide GC3 distribution, which significantly correlates with GC12. ENC-plot result—suggesting that most genes were close to the expected curve with only a few strayed away points. This indicates that mutational pressure and natural selection played an important role in the development of CUB. The Parity Rule 2 plot (PR2) result demonstrates that the usage of GC and AT was out of proportion. Interestingly, we identified 26 optimal codons in the G. duodenalis genome, ending with G or C. In addition, GC content, gene expression, and protein size also influence G. -

Ab253372 Β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit
Version 1 Last updated 14 June 2020 ab253372 β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit View β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit datasheet: www.abcam.com/ab253372 (use www.abcam.cn/ab253372 for China, or www.abcam.co.jp/ab253372 for Japan) For the measurement of β-Glucuronidase (GUS) activity in plant tissue. This product is for research use only and is not intended for diagnostic use. Copyright © 2020 Abcam. All rights reserved Table of Contents 1. Overview 1 2. Materials Supplied and Storage 2 3. Materials Required, Not Supplied 3 4. General guidelines, precautions, and troubleshooting 3 5. Reagent Preparation 4 6. Assay Procedure 5 7. Typical Data 7 8. Notes 9 Technical Support 10 Copyright © 2020 Abcam. All rights reserved 1. Overview Reporter genes are widely used as “markers” for analysis in gene regulation and localization, as well as for analysis of mutation altered genes. Expression of reporter genes can be measured by immunological assay, biochemical activity assay or by histochemical staining of cells or tissues. The β-glucuronidase (GUS) enzyme from E. coli (EC 3.2.1.31) has been well documented to provide desirable characteristics as a marker gene in transformed plants. The GUS reporter gene system has many advantages including stable expression of E. coli GUS enzyme, no interference with normal plant metabolism, and low intrinsic GUS activity in higher plants. The enzyme is also capable of tolerating amino-terminal additions, making it useful for study of plant organelle transport. Plants or other cell types are extracted with GUS extraction buffer. The extracted β-glucuronidase hydrolyzes the 4-MUG to the fluorescent compound 4-MU (pKa 8.2) and glucuronic acid. -

Pflash Phagemids Technical Manual
- Pg. 1 I. Description The pFlash™ phagemids are intendedfor the quantitative analysis of putativecis-elements and transacting factorsthat regulate eukaryoticgene regulation. The pFlash™ phagemids carry the coding region of the firefly (Photinuspyralis ) luciferase gene which is usedto monitor transcriptional activity in transfected eukaryotic cells. The assay of this reportergene is rapid, sensitive and quantitative and represents the efficiencyof transcription initiationutilizing the putative cis-elements cloned upstream/downstreamof the reporter gene. In addition the pFlash™ phagemids contain numerous features that facilitate the characterizationand mutagenesis of the putativecis-elements beingstudied. II. General Considerations A. Structure and Function There are three types of pFlash™ vectors: pFlash I™, pFlash II™, and SV40-pFlash™. Each pFlash™ phagemid carries the luciferase gene (luc) followedby the SV40 t intron containing the polyadenylation [poly(A)] signals which is the generic donor forthese signals and are used by every reporter gene vector that is currently available commercially. However the pFlash™ phagemids are distinguished by the fact that it is the only set in which the cryptic APl sequence (TGAGTCA) which is present in the wild type SV40 t intron has been specifically mutated by site-directed mutagenesis into an EcoRI (GAATTC) restriction site. The APl sequence is a strong eukaryotic enhancer element that is recognized by the ubiquitous Jun-Fos transcription factors as well as severalothers. The presence of this crypticenhancer in the t intronresults in spurious reporter gene expression and high background activity that has the potential to mask or otherwise interfere with the analysis of putative cis-elements that are being cloned upstream. The pFlash™ phagemids in which this cryptic site has been mutated are free of such activity and consistently exhibit extremely low signal-noise ratios. -

Molecular Mechanism of Light Responses in Neurospora: from Light-Induced Transcription to Photoadaptation
Downloaded from genesdev.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation Qiyang He and Yi Liu1 Department of Physiology, The University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA Blue light regulates many molecular and physiological activities in a large number of organisms. In Neurospora crassa, a eukaryotic model system for studying blue-light responses, the transcription factor and blue-light photoreceptor WHITE COLLAR-1 (WC-1) and its partner WC-2 are central to blue-light sensing. Neurospora’s light responses are transient, that is, following an initial acute phase of induction, light-regulated processes are down-regulated under continuous illumination, a phenomenon called photoadaptation. The molecular mechanism(s) of photoadaptation are not well understood. Here we show that a common mechanism controls the light-induced transcription of immediate early genes (such as frq, al-3, and vvd)inNeurospora, in which light induces the binding of identical large WC-1/WC-2 complexes (L-WCC) to the light response elements (LREs) in their promoters. Using recombinant proteins, we show that the WC complexes are functional without the requirement of additional factors. In vivo, WCC has a long period photocycle, indicating that it cannot be efficiently used for repeated light activation. Contrary to previous expectations, we demonstrate that the light-induced hyperphosphorylation of WC proteins inhibits bindings of the L-WCC to the LREs. We show that, in vivo, due to its rapid hyperphosphorylation, L-WCC can only bind transiently to LREs, indicating that WCC hyperphosphorylation is a critical process for photoadaptation.