Regulation of Gene Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporter Genes a Practical Guide M E T H O D S I N M O L E C U L a R B I O L O G Y™
Reporter Genes A Practical Guide M E T H O D S I N M O L E C U L A R B I O L O G Y™ John M. Walker, SERIES EDITOR 436. Avian Influenza Virus, edited by 413. Protein Structure Prediction, Second Edition, Erica Spackman, 2008 edited by Mohammed Zaki and Chris Bystroff, 435. Chromosomal Mutagenesis, edited by 2008 Greg Davis and Kevin J. Kayser, 2008 412. Neutrophil Methods and Protocols, edited by 434. Gene Therapy Protocols: Vol. 2: Design and Mark T. Quinn, Frank R. DeLeo, Characterization of Gene Transfer Vectors, and Gary M. Bokoch, 2007 edited by Joseph M. LeDoux, 2008 411. Reporter Genes: A Practical Guide, edited by 433. Gene Therapy Protocols: Vol. 1: Production Don Anson, 2007 and In Vivo Applications of Gene Transfer 410. Environmental Genomics, edited by Vectors, edited by Joseph M. LeDoux, 2008 Cristofre C. Martin, 2007 432. Organelle Proteomics, edited by 409. Immunoinformatics: Predicting Immunogenicity Delphine Pflieger and Jean Rossier, 2008 In Silico, edited by Darren R. Flower, 2007 431. Bacterial Pathogenesis: Methods and 408. Gene Function Analysis, edited by Protocols, edited by Frank DeLeo and Michael Ochs, 2007 Michael Otto, 2008 407. Stem Cell Assays, edited by Vemuri C. Mohan, 430. Hematopoietic Stem Cell Protocols, 2007 edited by Kevin D. Bunting, 2008 406. Plant Bioinformatics: Methods and Protocols, 429. Molecular Beacons: Signalling Nucleic Acid edited by David Edwards, 2007 Probes, Methods and Protocols, edited by 405. Telomerase Inhibition: Strategies and Andreas Marx and Oliver Seitz, 2008 Protocols, edited by Lucy Andrews and 428. Clinical Proteomics: Methods and Protocols, Trygve O. -

Construction of a Copper Bioreporter Screening, Characterization and Genetic Improvement of Copper-Sensitive Bacteria
Construction of a Copper Bioreporter Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath This thesis comprises 30 ECTS credits and is a compulsory part in the Master of Science With a Major in Industrial Biotechnology, 120 ECTS credits No. 9/2009 Construction of a Copper Bioreporter-Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath, [email protected] Master thesis Subject Category: Technology University College of Borås School of Engineering SE-501 90 BORÅS Telephone +46 033 435 4640 Examiner: Dr. Elisabeth Feuk-lagerstedt Supervisor, name: Dr. Saman Hosseinkhani Supervisor, address: Department of Biochemistry, Faculty of Biological Sciences, Tarbiat Modares University Tehran, Iran Date: 2009-12-14 Keywords: Copper Bioreporter, Luciferase assay, COP operon, pGL3, E. Coli BL-21 ii Dedicated to my parents to whom I owe and feel the deepest gratitude iii Acknowledgment I would like to express my best appreciation to my supervisor Dr. S. Hosseinkhani for technical supports, and my examiner Dr. E. Feuk-lagerstedt for her kind attention, also I want to thank Mr. A. Emamzadeh, Miss. M. Nazari, and other students of Biochemistry laboratory of Tarbiat Modares University for their kind corporations. iv Abstract In the nature, lots of organism applies different kinds of lights such as flourescence or luminescence for some purposes such as defense or hunting. Firefly luciferase and Bacterial luciferase are the most famous ones which have been used to design Biosensors or Bioreporters in recent decades. Their applications are so extensive from detecting pollutions in the environment to medical and treatment usages. -

Reporter Gene Assays
TCA-017 Reporter Gene Assays Abstract foreign to mammalian cells, and are biologically active without post-translational modification. Luminescent reporter gene assays provide a simple, fast, non-isotopic, and highly sensitive alternative to The high quantum efficiency of luciferase enables the popular radioactive chloramphenicol detection of less than 0.01 attomoles of luciferase, acetyltransferase (CAT) assay. This paper describes making it possible to scale down the assay to the a reporter gene assay in which cells were transfected 96-well microplate format.1,2 Although the luciferin/ with the firefly luciferase gene, which was quanti- luciferase reaction is often thought of as a flash fied using an enhanced flash luciferin substrate chemistry, recent advances have extended the pho- and the TopCount Microplate Scintillation and ton kinetics by the use of coenzyme A as a substrate Luminescence Counter. Co-transfection with the in the oxidation of luciferin.3 The resulting enhanced ß-galactosidase gene may be included to control for flash eliminates the need for elaborate reagent inter-assay transfection efficiency. Both of these injection devices. luminescent reporters can be quantified without reagent injection devices. The signal decays only slightly during the time required to read a microplate, Co-transfection with a second reporter gene is often and can be easily corrected for the decay by tandem performed to normalize transfection efficiency processing on TopCount. between assays. The ß-galactosidase gene is ideal for this application. Chemiluminescent substrates for ß- galactosidase are sensitive enough to allow detec- Introduction tion at low transfection efficiency, are available in compatible plasmid vectors, and are easily adaptable The use of reporter genes is a fundamental tool of 4 molecular biologists. -
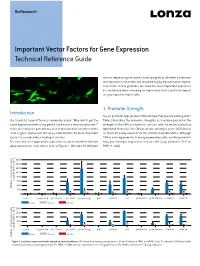
Important Vector Factors for Gene Expression Technical Reference Guide
BioResearch Important Vector Factors for Gene Expression Technical Reference Guide vectors expressing the same luciferase gene in different backbones and expression cassettes and obtained highly discriminative expres- sion levels. In this guideline, we show the most important aspects to be considered when choosing an expression vector and their impact on your experimental results. 1. Promoter Strength Introduction Is your promoter appropriate for the cell type that you are working with? Our Scientific Support Team is commonly asked: “Why don’t I get the Table 1 describes the promoter strengths as a relative percent of the same expression level of my gene if I use various vector backbones?” strength of the CMV promoter for various cells for which Lonza has There are many components to a vector that can have an effect on the Optimized Protocols. The CMV promoter activity is set to 100% based level of gene expression. Below you will find the 10 most important on their CAT assay values from the referenced publications. Although factors to consider when looking at vectors. CMV is a strong promoter in many mammalian cells, another promoter The selection of an appropriate expression vector is crucial for efficient may give stronger expression in your cells (e.g., promoter SV40 in gene expression. Just take a look at Figure 1. We tried 10 different BHK-21 cells). n 1600 1400 1200 1000 800 [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 600 400 Relative luciferase-expressio Relative 200 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT 4 hours 24 hours 48 hours n 2000 1500 1000 500 0 Control Program Only pGL3-Control pGL3-CMV pmax Cloning™ pSG5 pcDNA3.1 pFP pIRES-MCS B pcDNA4 HisMax pCMVbeta pCMV-TNT [% of pGL3-CMV at 24 h] 24 at pGL3-CMV [% of 4 hours 24 hours 48 hours Relative luciferase-expressio Relative Figure 1: Luciferase expression levels depend on vector backbones. -

Robust Normalization of Luciferase Reporter Data
Technical Note Robust Normalization of Luciferase Reporter Data Andrea Repele † and Manu * Department of Biology, University of North Dakota, Grand Forks, ND 58202, USA * Correspondence: [email protected] † Current address: JMB—Center for Immunity and Immunotherapies, Seattle Children’s Hospital, Seattle, WA 98105, USA. Received: 17 June 2019; Accepted: 22 July 2019; Published: 25 July 2019 Abstract: Transient Luciferase reporter assays are widely used in the study of gene regulation and intracellular cell signaling. In order to control for sample-to-sample variation in luminescence arising from variability in transfection efficiency and other sources, an internal control reporter is co-transfected with the experimental reporter. The luminescence of the experimental reporter is normalized against the control by taking the ratio of the two. Here we show that this method of normalization, “ratiometric”, performs poorly when the transfection efficiency is low and leads to biased estimates of relative activity. We propose an alternative methodology based on linear regression that is much better suited for the normalization of reporter data, especially when transfection efficiency is low. We compare the ratiometric method against three regression methods on both simulated and empirical data. Our results suggest that robust errors-in-variables (REIV) regression performs the best in normalizing Luciferase reporter data. We have made the R code for Luciferase data normalization using REIV available on GitHub. Keywords: luciferase reporter; transfection efficiency; normalization; gene regulation; promoter; enhancer 1. Introduction Transient reporter assays are an important and widely used tool in the study of gene regulation [1–4], intracellular cell signaling [5–7], and other areas of molecular, cellular, and developmental biology [8–10]. -

I = Chpt 15. Positive and Negative Transcriptional Control at Lac BMB
BMB 400 Part Four - I = Chpt 15. Positive and Negative Transcriptional Control at lac B M B 400 Part Four: Gene Regulation Section I = Chapter 15 POSITIVE AND NEGATIVE CONTROL SHOWN BY THE lac OPERON OF E. COLI A. Definitions and general comments 1. Operons An operon is a cluster of coordinately regulated genes. It includes structural genes (generally encoding enzymes), regulatory genes (encoding, e.g. activators or repressors) and regulatory sites (such as promoters and operators). 2. Negative versus positive control a. The type of control is defined by the response of the operon when no regulatory protein is present. b. In the case of negative control, the genes in the operon are expressed unless they are switched off by a repressor protein. Thus the operon will be turned on constitutively (the genes will be expressed) when the repressor in inactivated. c. In the case of positive control, the genes are expressed only when an active regulator protein, e.g. an activator, is present. Thus the operon will be turned off when the positive regulatory protein is absent or inactivated. Table 4.1.1. Positive vs. negative control BMB 400 Part Four - I = Chpt 15. Positive and Negative Transcriptional Control at lac 3. Catabolic versus biosynthetic operons a. Catabolic pathways catalyze the breakdown of nutrients (the substrate for the pathway) to generate energy, or more precisely ATP, the energy currency of the cell. In the absence of the substrate, there is no reason for the catabolic enzymes to be present, and the operon encoding them is repressed. In the presence of the substrate, when the enzymes are needed, the operon is induced or de-repressed. -

Ab253372 Β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit
Version 1 Last updated 14 June 2020 ab253372 β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit View β-Glucuronidase (GUS) Reporter Gene Activity Detection Kit datasheet: www.abcam.com/ab253372 (use www.abcam.cn/ab253372 for China, or www.abcam.co.jp/ab253372 for Japan) For the measurement of β-Glucuronidase (GUS) activity in plant tissue. This product is for research use only and is not intended for diagnostic use. Copyright © 2020 Abcam. All rights reserved Table of Contents 1. Overview 1 2. Materials Supplied and Storage 2 3. Materials Required, Not Supplied 3 4. General guidelines, precautions, and troubleshooting 3 5. Reagent Preparation 4 6. Assay Procedure 5 7. Typical Data 7 8. Notes 9 Technical Support 10 Copyright © 2020 Abcam. All rights reserved 1. Overview Reporter genes are widely used as “markers” for analysis in gene regulation and localization, as well as for analysis of mutation altered genes. Expression of reporter genes can be measured by immunological assay, biochemical activity assay or by histochemical staining of cells or tissues. The β-glucuronidase (GUS) enzyme from E. coli (EC 3.2.1.31) has been well documented to provide desirable characteristics as a marker gene in transformed plants. The GUS reporter gene system has many advantages including stable expression of E. coli GUS enzyme, no interference with normal plant metabolism, and low intrinsic GUS activity in higher plants. The enzyme is also capable of tolerating amino-terminal additions, making it useful for study of plant organelle transport. Plants or other cell types are extracted with GUS extraction buffer. The extracted β-glucuronidase hydrolyzes the 4-MUG to the fluorescent compound 4-MU (pKa 8.2) and glucuronic acid. -

Pflash Phagemids Technical Manual
- Pg. 1 I. Description The pFlash™ phagemids are intendedfor the quantitative analysis of putativecis-elements and transacting factorsthat regulate eukaryoticgene regulation. The pFlash™ phagemids carry the coding region of the firefly (Photinuspyralis ) luciferase gene which is usedto monitor transcriptional activity in transfected eukaryotic cells. The assay of this reportergene is rapid, sensitive and quantitative and represents the efficiencyof transcription initiationutilizing the putative cis-elements cloned upstream/downstreamof the reporter gene. In addition the pFlash™ phagemids contain numerous features that facilitate the characterizationand mutagenesis of the putativecis-elements beingstudied. II. General Considerations A. Structure and Function There are three types of pFlash™ vectors: pFlash I™, pFlash II™, and SV40-pFlash™. Each pFlash™ phagemid carries the luciferase gene (luc) followedby the SV40 t intron containing the polyadenylation [poly(A)] signals which is the generic donor forthese signals and are used by every reporter gene vector that is currently available commercially. However the pFlash™ phagemids are distinguished by the fact that it is the only set in which the cryptic APl sequence (TGAGTCA) which is present in the wild type SV40 t intron has been specifically mutated by site-directed mutagenesis into an EcoRI (GAATTC) restriction site. The APl sequence is a strong eukaryotic enhancer element that is recognized by the ubiquitous Jun-Fos transcription factors as well as severalothers. The presence of this crypticenhancer in the t intronresults in spurious reporter gene expression and high background activity that has the potential to mask or otherwise interfere with the analysis of putative cis-elements that are being cloned upstream. The pFlash™ phagemids in which this cryptic site has been mutated are free of such activity and consistently exhibit extremely low signal-noise ratios. -

Bioluminescent Reporter Gene Assays Protocols and Applications
|||||||| 8Bioluminescent Reporter Gene Assays PR CONTENTS I. Introduction 1 O A. Luminescence versus Fluorescence 1 B. Bioluminescent Reporters 2 T OCOLS C. Applications 3 II. Luciferase Genes and Vectors 4 A. Biology and Enzymology 4 B. Gene Optimization 6 C. pGL4 Luciferase Reporter Vectors 8 D. pNL Vectors 8 & III. Luciferase Reporter Assays and Protocols 8 A. Single-Reporter Assays 9 APPLIC B. Dual-Reporter Assays 10 C. Live-Cell Substrates 12 D. Bioluminescent Reporters to Normalize for Changes in Cell Physiology 12 E. Bioluminescent Reporters to Monitor RNA Interference 13 A F. Bioluminescent Reporters in Cell-Signaling Assays 13 TIONS G. Luciferase Reporter Cell Lines 15 H. Bioluminescent Reporters to Study Protein Dynamics 15 I. Promega Reporter Vectors and Assays 16 IV. Getting the Most Out of Your Genetic Reporter GUIDE Assays 16 A. Introduction 16 B. Choice of Reporter Gene and Assay 16 C. Reporter Construct Design 16 D. Controls 17 E. Transfection Parameters 17 F. Assay Timing 17 G. Cells and Cell Culture Conditions 18 H. Special Considerations for High-Throughput Assays 18 V. References 19 VI. Vector Tables 20 Protocols & Applications Guide www.promega.com rev. 4/15 |||||||| 8Bioluminescent Reporter Gene Assays PR I. Introduction Fundamentally, a reporter assay is a means to translate a biomolecular effect into an observable parameter. While Genetic reporter systems have contributed greatly to the there are theoretically many strategies by which this can study of eukaryotic gene expression and regulation. O be achieved, in practice the reporter assays capable of Although reporter genes have played a significant role in delivering the speed, accuracy and sensitivity necessary T numerous applications, both in vitro and in vivo, they are for effective screening are based on photon production. -

Pgl3 Luciferase Reporter Vectors Technical Manual TM033
TECHNICAL MANUAL pGL3 Luciferase Reporter Vectors Instructions For Use of Products E1741, E1751, E1761 and E1771 Revised 6/15 TM033 pGL3 Luciferase Reporter Vectors All technical literature is available at: www.promega.com/protocols/ Visit the web site to verify that you are using the most current version of this Technical Manual. E-mail Promega Technical Services if you have questions on use of this system: [email protected] 1. Description .........................................................................................................................................2 2. Product Components and Storage Conditions ........................................................................................2 3. pGL3 Vector Maps and Sequence Reference Points ................................................................................2 3.A. pGL3-Basic Vector ......................................................................................................................3 3.B. pGL3-Enhancer Vector ................................................................................................................4 3.C. pGL3-Promoter Vector ................................................................................................................5 3.D. pGL3-Control Vector ..................................................................................................................6 4. Cloning Methods .................................................................................................................................7 -

Reporter Gene
Reporter gene Programme: B.Sc (H) Botany Course Title: Plant Biotechnology Course code: BOTY 3014 Prof. Shahana Majumder Department of Botany Mahatma Gandhi Central University, Motihari Disclaimer • These materials are taken/borrowed/modified/compiled from various sources like research articles and freely available internet websites, and are meant to be used solely for the teaching purpose in a public university, and solely for the use of UG students. Selection of a transgene • The use of a marker gene in a transformation process aims to select transformed cells that is expressing the gene of Interest. • These selectable marker genes are often known as Reporter gene • Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured. Marker genes for selection enable the identification and selection of genetically modified cells. http://www.genopomii.un ina.it/genohort/files/Lezio ne_15_Plant%20cell%20s uspensions.pdf Tissue culture Treat with appropriate method to detect the Explant transgenic cells Agro inoculation Genetically modified cells • To introduce a reporter gene into an organism, scientists place the reporter gene and the gene of interest under the control of same promoter in the same DNA construct . • This typically involves removing the stop codon from a cDNA sequence coding for the first protein, then appending the cDNA sequence of the second protein in frame through ligation . Spacer Gene 1 Protein 2 Spacer Protein 1 http://2010.igem.org/Team:BIOTEC_Dresden/Fusion_Protein • Here often a linker (or "spacer") peptides are also added, which make it more likely that the proteins fold independently and behave as independent proteins • It is important to use a reporter gene that is not natively expressed in the cell or organism under study, since the expression of the reporter is being used as a marker for successful uptake of the gene of interest. -

How We Can Use Reporter Genes to Light up the Cellular World by Melanie Rose
Now you see me: How we can use reporter genes to light up the cellular world By Melanie Rose Jellyfish, fireflies and mushrooms: What do they all have in common? They come from different environments and have wildly different lifecycles, but they all have a secret hidden away where nobody can see it. That is, until they want you to. Light! Bioluminescence, to be exact, or the production of light by living organisms! Some members of these groups are able to produce light through a chemical reaction, while others simply produce a protein that glows on its own. Scientists have been quick to harness these unique properties for their own research purposes. When bioluminescence is taken and used in another organism it is referred to as a reporter gene. Reporter genes are valuable tools as they can illuminate cells and allow for visual assessments such as cell tracking or morphological analysis. Similar to bioluminescence, fluorescent proteins can serve as useful indicators of gene expression and protein distribution in living cells. With the development and discovery of multiple fluorescent proteins from many organisms there has been a change in how cellular functions are being studied. The most commonly used reporter is a fluorescent protein which glows green when exposed to a specific wavelength of light. This reporter is called green fluorescent protein, or GFP. The protein originally came from the jellyfish Aequorea victoria, however, researchers have artificially reproduced it in the lab with enhanced brightness and function. GFP has been joined with many proteins/ genes of interest without hindering how the protein functions.