A Firefly Luciferase Dual Color Bioluminescence Reporter Assay
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Immobilization of Firefly Luciferase on PVA-Co-PE Nanofibers Membrane
Research Article www.acsami.org Immobilization of Firefly Luciferase on PVA-co-PE Nanofibers Membrane as Biosensor for Bioluminescent Detection of ATP † † Wenwen Wang, Qinghua Zhao, Mengying Luo, Mufang Li, Dong Wang,* Yuedan Wang, and Qiongzhen Liu School of Materials Science and Engineering, Wuhan Textile University, Wuhan 430073, China ABSTRACT: The bioluminescent reaction catalyzed by firefly luciferase has become widely established as an outstanding analytical system for assay of adenosine triphosphate (ATP). When in solution, the luciferase is unstable and cannot be reused. The problem can be partially solved by immobilizing the luciferase on solid substrates. The poly(vinyl alcohol-co-ethylene) (PVA-co-PE) nanofibers membrane has abundant active hydroxyl groups on the surface. The PVA-co-PE nanofibers membrane was first activated by cyanuric chloride with triazinyl group. Then the activated PVA-co- PE nanofibers membrane was subsequently reacted with 1,3-propanediamine and biotin. The firefly luciferase was immobilized onto the surface of 1,3-propanediamine- and biotin-functionalized membranes. The surface chemical structure and morphologies of nanofibers membranes were characterized by FTIR-ATR spectra and SEM. The hydrophilicity of membranes was tested by water contact angle measurements. The detection of fluorescence intensity displayed that the firefly-luciferase-immobilized PVA-co-PE nanofibers membranes indicated high catalytic activity and efficiency. Especially, the firefly-luciferase-immobilized nanofiber membrane which was functionalized -

Reporter Genes a Practical Guide M E T H O D S I N M O L E C U L a R B I O L O G Y™
Reporter Genes A Practical Guide M E T H O D S I N M O L E C U L A R B I O L O G Y™ John M. Walker, SERIES EDITOR 436. Avian Influenza Virus, edited by 413. Protein Structure Prediction, Second Edition, Erica Spackman, 2008 edited by Mohammed Zaki and Chris Bystroff, 435. Chromosomal Mutagenesis, edited by 2008 Greg Davis and Kevin J. Kayser, 2008 412. Neutrophil Methods and Protocols, edited by 434. Gene Therapy Protocols: Vol. 2: Design and Mark T. Quinn, Frank R. DeLeo, Characterization of Gene Transfer Vectors, and Gary M. Bokoch, 2007 edited by Joseph M. LeDoux, 2008 411. Reporter Genes: A Practical Guide, edited by 433. Gene Therapy Protocols: Vol. 1: Production Don Anson, 2007 and In Vivo Applications of Gene Transfer 410. Environmental Genomics, edited by Vectors, edited by Joseph M. LeDoux, 2008 Cristofre C. Martin, 2007 432. Organelle Proteomics, edited by 409. Immunoinformatics: Predicting Immunogenicity Delphine Pflieger and Jean Rossier, 2008 In Silico, edited by Darren R. Flower, 2007 431. Bacterial Pathogenesis: Methods and 408. Gene Function Analysis, edited by Protocols, edited by Frank DeLeo and Michael Ochs, 2007 Michael Otto, 2008 407. Stem Cell Assays, edited by Vemuri C. Mohan, 430. Hematopoietic Stem Cell Protocols, 2007 edited by Kevin D. Bunting, 2008 406. Plant Bioinformatics: Methods and Protocols, 429. Molecular Beacons: Signalling Nucleic Acid edited by David Edwards, 2007 Probes, Methods and Protocols, edited by 405. Telomerase Inhibition: Strategies and Andreas Marx and Oliver Seitz, 2008 Protocols, edited by Lucy Andrews and 428. Clinical Proteomics: Methods and Protocols, Trygve O. -

Bioluminescent Properties of Semi-Synthetic Obelin and Aequorin Activated by Coelenterazine Analogues with Modifications of C-2, C-6, and C-8 Substituents
International Journal of Molecular Sciences Article Bioluminescent Properties of Semi-Synthetic Obelin and Aequorin Activated by Coelenterazine Analogues with Modifications of C-2, C-6, and C-8 Substituents 1, 2,3, 1 2, Elena V. Eremeeva y , Tianyu Jiang y , Natalia P. Malikova , Minyong Li * and Eugene S. Vysotski 1,* 1 Photobiology Laboratory, Institute of Biophysics SB RAS, Federal Research Center “Krasnoyarsk Science Center SB RAS”, Krasnoyarsk 660036, Russia; [email protected] (E.V.E.); [email protected] (N.P.M.) 2 Key Laboratory of Chemical Biology (MOE), Department of Medicinal Chemistry, School of Pharmaceutical Sciences, Shandong University, Jinan 250012, China; [email protected] 3 State Key Laboratory of Microbial Technology, Shandong University–Helmholtz Institute of Biotechnology, Shandong University, Qingdao 266237, China * Correspondence: [email protected] (M.L.); [email protected] (E.S.V.); Tel.: +86-531-8838-2076 (M.L.); +7-(391)-249-44-30 (E.S.V.); Fax: +86-531-8838-2076 (M.L.); +7-(391)-290-54-90 (E.S.V.) These authors contributed equally to this work. y Received: 23 June 2020; Accepted: 27 July 2020; Published: 30 July 2020 Abstract: Ca2+-regulated photoproteins responsible for bioluminescence of a variety of marine organisms are single-chain globular proteins within the inner cavity of which the oxygenated coelenterazine, 2-hydroperoxycoelenterazine, is tightly bound. Alongside with native coelenterazine, photoproteins can also use its synthetic analogues as substrates to produce flash-type bioluminescence. However, information on the effect of modifications of various groups of coelenterazine and amino acid environment of the protein active site on the bioluminescent properties of the corresponding semi-synthetic photoproteins is fragmentary and often controversial. -

Crystal Structure of Firefly Luciferase Throws Light on a Superfamily of Adenylate-Forming Enzymes Elena Conti, Nick P Franks and Peter Brick*
Research Article 287 Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes Elena Conti, Nick P Franks and Peter Brick* Background: Firefly luciferase is a 62 kDa protein that catalyzes the production Address: Biophysics Section, Blackett Laboratory, of light. In the presence of MgATP and molecular oxygen, the enzyme oxidizes its Imperial College, London SW7 2BZ, UK. substrate, firefly luciferin, emitting yellow-green light. The reaction proceeds *Corresponding author. through activation of the substrate to form an adenylate intermediate. Firefly luciferase shows extensive sequence homology with a number of enzymes that Key words: acyl-coenzyme A ligase, adenylate, utilize ATP in adenylation reactions. firefly luciferase, peptide synthetase, X-ray crystallography Results: We have determined the crystal structure of firefly luciferase at 2.0 Å Received: 30 Nov 1995 resolution. The protein is folded into two compact domains. The large N-terminal Revisions requested: 21 Dec 1995 domain consists of a b-barrel and two b-sheets. The sheets are flanked by Revisions received: 15 Jan 1996 a-helices to form an ababa five-layered structure. The C-terminal portion of the Accepted: 31 Jan 1996 molecule forms a distinct domain, which is separated from the N-terminal domain Structure 15 March 1996, 4:287–298 by a wide cleft. © Current Biology Ltd ISSN 0969-2126 Conclusions: Firefly luciferase is the first member of a superfamily of homologous enzymes, which includes acyl-coenzyme A ligases and peptide synthetases, to have its structure characterized. The residues conserved within the superfamily are located on the surfaces of the two domains on either side of the cleft, but are too far apart to interact simultaneously with the substrates. -

Discovery and Protein Engineering of Baeyer-Villiger Monooxygenases
Discovery and Protein Engineering of Baeyer-Villiger monooxygenases Inauguraldissertation zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften (Dr. rer. nat.) der Mathematisch-Naturwissenschaftlichen Fakultät der Ernst-Moritz-Arndt-Universität Greifswald vorgelegt von Andy Beier geboren am 11.10.1988 in Parchim Greifswald, den 02.08.2017 I Dekan: Prof. Dr. Werner Weitschies 1. Gutachter: Prof. Dr. Uwe T. Bornscheuer 2. Gutachter: Prof. Dr. Marko Mihovilovic Tag der Promotion: 24.10.2017 II We need to learn to want what we have, not to have what we want, in order to get stable and steady happiness. - The Dalai Lama - III List of abbreviations % Percent MPS Methyl phenyl sulfide % (v/v) % volume per volume MPSO Methyl phenyl sulfoxide % (w/v) % weight per volume MPSO2 Methyl phenyl sulfone °C Degrees Celsius MTS Methyl p-tolyl sulfide µM µmol/L MTSO Methyl p-tolyl sulfoxide aa Amino acids MTSO2 Methyl p-tolyl sulfone + AGE Agarose gel electrophoresis NAD Nicotinamide adenine dinucleotide, oxidized aq. dest. Distilled water NADH Nicotinamide adenine dinucleotide, reduced + BLAST Basic Local Alignment Search NADP Nicotinamide adenine dinucleotide Tool phosphate, oxidized bp Base pair(s) NADPH Nicotinamide adenine dinucleotide phosphate, reduced BVMO Baeyer-Villiger monooxyge- OD600 Optical density at 600 nm nase CHMO Cyclohexanone monooxyge- PAGE Polyacrylamide gel electrophoresis nase Da Dalton PAMO Phenylacetone monooxygenase DMF Dimethyl formamide PCR Polymerase chain reaction DMSO Dimethyl sulfoxide PDB Protein Data Bank DMSO2 Dimethyl sulfone rpm Revolutions per minute DNA Desoxyribonucleic acid rv Reverse dNTP Desoxynucleoside triphosphate SDS Sodium dodecyl sulfate E. coli Escherichia coli SOC Super Optimal broth with Catabolite repression ee Enantiomeric excess TAE TRIS-Acetate-EDTA FAD Flavin adenine dinucleotide TB Terrific broth Fig. -

Cloning of Firefly Luciferase Cdna and the Expression of Active
Proc. Natl. Acad. Sci. USA Vol. 82, pp. 7870-7873, December 1985 Biochemistry Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli (bioluminescence/Photinus pyralis/antibody screening/expression vector/recombinant DNA) JEFFREY R. DE WET*, KEITH V. WOODt, DONALD R. HELINSKI*, AND MARLENE DELUCAt Departments of *Biology and tChemistry, University of California, San Diego, La Jolla, CA 92093 Communicated by W. D. McElroy, July 26, 1985 ABSTRACT A cDNA library was constructed from firefly library was screened with anti-P. pyralis luciferase antibody, (Photinuspyralis) lantern poly(A)I RNA, using the Escherichia using a chromogenic detection technique (8), and several coli expression vector Xgtll. The library was screened with cDNA clones were isolated and characterized. These clones anti-P. pyralis luciferase (Photinus luciferin:oxygen 4-oxidore- were found to be homologous to the mRNA that encodes ductase, EC 1.13.12.7) antibody, and several cDNA clones luciferase. The largest luciferase cDNA clone that was expressing luciferase antigens were isolated. One clone, ALucl, isolated was able to direct the synthesis of active luciferase contained 1.5 kilobase pairs of cDNA that hybridized to a 1.9- in E. coli. to 2.0-kilobase band on a nitrocellulose blot of electrophoreti- cally fractionated lantern RNA. Hybridization of the cloned MATERIALS AND METHODS cDNA to lantern poly(A)I RNA selected an RNA that directed the in vitro synthesis of a single polypeptide. This polypeptide Enzymes and Strains. Restriction endonucleases and E. coli comigrated with luciferase on NaDodSO4/PAGE and produced DNA polymerase I were purchased from New England bioluminescence upon the addition of luciferin and ATP. -

Bioluminescence Is Produced by a Firefly-Like Luciferase but an Entirely
www.nature.com/scientificreports OPEN New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a Received: 8 November 2017 Accepted: 1 February 2018 frefy-like luciferase but an entirely Published: xx xx xxxx new luciferin Oliver C. Watkins1,2, Miriam L. Sharpe 1, Nigel B. Perry 2 & Kurt L. Krause 1 The New Zealand glowworm, Arachnocampa luminosa, is well-known for displays of blue-green bioluminescence, but details of its bioluminescent chemistry have been elusive. The glowworm is evolutionarily distant from other bioluminescent creatures studied in detail, including the frefy. We have isolated and characterised the molecular components of the glowworm luciferase-luciferin system using chromatography, mass spectrometry and 1H NMR spectroscopy. The purifed luciferase enzyme is in the same protein family as frefy luciferase (31% sequence identity). However, the luciferin substrate of this enzyme is produced from xanthurenic acid and tyrosine, and is entirely diferent to that of the frefy and known luciferins of other glowing creatures. A candidate luciferin structure is proposed, which needs to be confrmed by chemical synthesis and bioluminescence assays. These fndings show that luciferases can evolve independently from the same family of enzymes to produce light using structurally diferent luciferins. Glowworms are found in New Zealand and Australia, and are a major tourist attraction at sites located across both countries. In contrast to luminescent beetles such as the frefy (Coleoptera), whose bioluminescence has been well characterised (reviewed by ref.1), the molecular details of glowworm bioluminescence have remained elusive. Tese glowworms are the larvae of fungus gnats of the genus Arachnocampa, with eight species endemic to Australia and a single species found only in New Zealand2. -

Construction of a Copper Bioreporter Screening, Characterization and Genetic Improvement of Copper-Sensitive Bacteria
Construction of a Copper Bioreporter Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath This thesis comprises 30 ECTS credits and is a compulsory part in the Master of Science With a Major in Industrial Biotechnology, 120 ECTS credits No. 9/2009 Construction of a Copper Bioreporter-Screening, characterization and genetic improvement of copper-sensitive bacteria Puria Motamed Fath, [email protected] Master thesis Subject Category: Technology University College of Borås School of Engineering SE-501 90 BORÅS Telephone +46 033 435 4640 Examiner: Dr. Elisabeth Feuk-lagerstedt Supervisor, name: Dr. Saman Hosseinkhani Supervisor, address: Department of Biochemistry, Faculty of Biological Sciences, Tarbiat Modares University Tehran, Iran Date: 2009-12-14 Keywords: Copper Bioreporter, Luciferase assay, COP operon, pGL3, E. Coli BL-21 ii Dedicated to my parents to whom I owe and feel the deepest gratitude iii Acknowledgment I would like to express my best appreciation to my supervisor Dr. S. Hosseinkhani for technical supports, and my examiner Dr. E. Feuk-lagerstedt for her kind attention, also I want to thank Mr. A. Emamzadeh, Miss. M. Nazari, and other students of Biochemistry laboratory of Tarbiat Modares University for their kind corporations. iv Abstract In the nature, lots of organism applies different kinds of lights such as flourescence or luminescence for some purposes such as defense or hunting. Firefly luciferase and Bacterial luciferase are the most famous ones which have been used to design Biosensors or Bioreporters in recent decades. Their applications are so extensive from detecting pollutions in the environment to medical and treatment usages. -

Muscle Regeneration Controlled by a Designated DNA Dioxygenase
Wang et al. Cell Death and Disease (2021) 12:535 https://doi.org/10.1038/s41419-021-03817-2 Cell Death & Disease ARTICLE Open Access Muscle regeneration controlled by a designated DNA dioxygenase Hongye Wang1, Yile Huang2,MingYu3,YangYu1, Sheng Li4, Huating Wang2,5,HaoSun2,5,BingLi 3, Guoliang Xu6,7 andPingHu4,8,9 Abstract Tet dioxygenases are responsible for the active DNA demethylation. The functions of Tet proteins in muscle regeneration have not been well characterized. Here we find that Tet2, but not Tet1 and Tet3, is specifically required for muscle regeneration in vivo. Loss of Tet2 leads to severe muscle regeneration defects. Further analysis indicates that Tet2 regulates myoblast differentiation and fusion. Tet2 activates transcription of the key differentiation modulator Myogenin (MyoG) by actively demethylating its enhancer region. Re-expressing of MyoG in Tet2 KO myoblasts rescues the differentiation and fusion defects. Further mechanistic analysis reveals that Tet2 enhances MyoD binding by demethylating the flanking CpG sites of E boxes to facilitate the recruitment of active histone modifications and increase chromatin accessibility and activate its transcription. These findings shed new lights on DNA methylation and pioneer transcription factor activity regulation. Introduction Ten-Eleven Translocation (Tet) family of DNA dioxy- 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Skeletal muscles can regenerate due to the existence of genases catalyze the active DNA demethylation and play muscle stem cells (MuSCs)1,2. The normally quiescent critical roles in embryonic development, neural regen- MuSCs are activated after muscle injury and further dif- eration, oncogenesis, aging, and many other important – ferentiate to support muscle regeneration3,4. -

Food Derived from Herbicide-Tolerant Soybean Line SYHT0H2 Summary
Supporting document 1 Safety assessment – Application A1081 Food derived from Herbicide-tolerant Soybean Line SYHT0H2 Summary and Conclusions Background A genetically modified (GM) soybean line with OECD Unique Identifier SYN-0000H2-5, hereafter referred to as soybean SYHT0H2, has been developed to be tolerant to two herbicides with different modes of action, namely glufosinate-ammonium and mesotrione. Tolerance to glufosinate ammonium is achieved through expression of the enzyme phosphinothricin acetyltransferase (PAT). PAT is encoded by a pat gene obtained from the soil bacterium Streptomyces viridochromogenes. Tolerance to mesotrione is achieved through expression of the AvHPPD-03 protein encoded by the avhppd-03 gene from oat (Avena sativa). In conducting a safety assessment of food derived from soybean line SYHT0H2, a number of criteria have been addressed including: a characterisation of the transferred gene and its origin, function and stability in the soybean genome; the changes at the level of DNA, protein and in the whole food; compositional analyses; evaluation of intended and unintended changes; and the potential for the newly expressed proteins to be either allergenic or toxic in humans. This safety assessment report addresses only food safety and nutritional issues. It therefore does not address: environmental risks related to the environmental release of GM plants used in food production the safety of animal feed or animals fed with feed derived from GM plants the safety of food derived from the non-GM (conventional) plant. History of Use Soybean (Glycine max) is grown as a commercial crop in over 35 countries worldwide. Soybean-derived products have a range of food and feed as well as industrial uses and have a long history of safe use for both humans and livestock. -
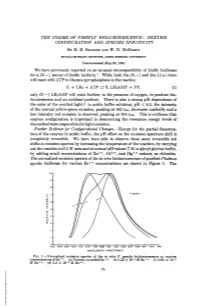
The Colors of Firefly Bioluminescence: Enzyme Configuration and Species Specificity by H
THE COLORS OF FIREFLY BIOLUMINESCENCE: ENZYME CONFIGURATION AND SPECIES SPECIFICITY BY H. H. SELIGER AND W. D. MCELROY MCCOLLUM-PRATT INSTITUTE, JOHNS HOPKINS UNIVERSITY Communicated May 25, 1964 We have previously reported on an unusual stereospecificity of firefly luciferase for a D(-) isomer of firefly luciferin.' While both the D(-) and the L(+) form will react with ATP to liberate pyrophosphate in the reaction E + LH2 + ATP =- E. LH2AMP + PP, (1) only D(-) LH2AMP will react further, in the presence of oxygen, to produce bio- luminescence and an oxidized product. There is also a strong pH dependence of the color of the emitted light;2 in acidic buffer solutions, pH < 6.5, the intensity of the normal yellow-green emission, peaking at 562 ml,, decreases markedly and a low intensity red emission is observed, peaking at 616 miu. This is evidence that enzyme configuration is important in determining the resonance energy levels of the excited state responsible for light emission. Further Evidence for Configurational Changes.-Except for the partial denatura- tion of the enzyme in acidic buffer, the pH effect on the emission spectrum shift is completely reversible. We have been able to observe these same reversible red shifts in emission spectra by increasing the temperature of the reaction, by carrying out the reaction in 0.2 M urea and at normal pH values (7.6) in glycyl glycine buffer, by adding small concentrations of Zn++, Cd++, and Hg++ cations, as chlorides. The normalized emission spectra of the in vitro bioluminescence of purified Photinus pyralis luciferase for various Zn++ concentrations are shown in Figure 1. -

Relating Metatranscriptomic Profiles to the Micropollutant
1 Relating Metatranscriptomic Profiles to the 2 Micropollutant Biotransformation Potential of 3 Complex Microbial Communities 4 5 Supporting Information 6 7 Stefan Achermann,1,2 Cresten B. Mansfeldt,1 Marcel Müller,1,3 David R. Johnson,1 Kathrin 8 Fenner*,1,2,4 9 1Eawag, Swiss Federal Institute of Aquatic Science and Technology, 8600 Dübendorf, 10 Switzerland. 2Institute of Biogeochemistry and Pollutant Dynamics, ETH Zürich, 8092 11 Zürich, Switzerland. 3Institute of Atmospheric and Climate Science, ETH Zürich, 8092 12 Zürich, Switzerland. 4Department of Chemistry, University of Zürich, 8057 Zürich, 13 Switzerland. 14 *Corresponding author (email: [email protected] ) 15 S.A and C.B.M contributed equally to this work. 16 17 18 19 20 21 This supporting information (SI) is organized in 4 sections (S1-S4) with a total of 10 pages and 22 comprises 7 figures (Figure S1-S7) and 4 tables (Table S1-S4). 23 24 25 S1 26 S1 Data normalization 27 28 29 30 Figure S1. Relative fractions of gene transcripts originating from eukaryotes and bacteria. 31 32 33 Table S1. Relative standard deviation (RSD) for commonly used reference genes across all 34 samples (n=12). EC number mean fraction bacteria (%) RSD (%) RSD bacteria (%) RSD eukaryotes (%) 2.7.7.6 (RNAP) 80 16 6 nda 5.99.1.2 (DNA topoisomerase) 90 11 9 nda 5.99.1.3 (DNA gyrase) 92 16 10 nda 1.2.1.12 (GAPDH) 37 39 6 32 35 and indicates not determined. 36 37 38 39 S2 40 S2 Nitrile hydration 41 42 43 44 Figure S2: Pearson correlation coefficients r for rate constants of bromoxynil and acetamiprid with 45 gene transcripts of ECs describing nucleophilic reactions of water with nitriles.