Joha Rice: an Aromatic Indigenous Rice of Assam, India Contains Flavanoids and Phenolic Substances and Shows Good Antioxidant Activities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Trait Expression Studies of Indigenous Joha Rice of Assam Under Organic
Journal of Pharmacognosy and Phytochemistry 2019; 8(3): 4290-4295 E-ISSN: 2278-4136 P-ISSN: 2349-8234 JPP 2019; 8(3): 4290-4295 Trait expression studies of indigenous Joha rice of Received: 07-03-2019 Accepted: 09-04-2019 Assam under organic and inorganic culture Khirud Panging Department of Plant Breeding Khirud Panging, Debojit Sarma, Rumjum Goswami Phukon and and Genetics, Assam Shantanu Das Agricultural University, Jorhat, Assam, India Abstract Debojit Sarma The present investigation was conducted to evaluate 12 indigenous Joha rice genotypes for different yield Department of Plant Breeding characters under inorganic and organic cultures. The materials were characterized during sali season of and Genetics, Assam 2015-16 at the Instruction-cum-Research (ICR), Assam Agricultural University (AAU), Jorhat using Agricultural University, Jorhat, Randomized Block Design (RBD) with three replications. Analysis of variance revealed the existence of Assam, India significant differences among the genotypes for all the traits under both inorganic and organic cultures. Among all the characters, panicle weight, grains/panicle, 1000-grain weight, grain yield/plant, straw Rumjum Goswami Phukon yield/plant and harvest index exhibited moderate to high estimates of the genotypic coefficient of Department of Plant Breeding variation (GCV) and phenotypic coefficient of variation (PCV) were recorded under organic and and Genetics, Assam inorganic culture. Also, high estimates of heritability were recorded for days to first flowering, days to Agricultural University, Jorhat, Assam, India 50% flowering, plant height, grains/panicle, 1000-grain weight, and harvest index. High heritability coupled with high genetic advance was observed only for 1000-grain weight under both the cultures. -

Agricultural and Food
REGISTERED GEOGRAPHICAL INDICATIONS INDIA- AGRICULTURAL AND FOOD S. Application Geographical Goods (As State No No. Indications per Sec 2 (f) of GIG Act 1999 ) 1 143 Guntur Sannam Chilli Agricultural Andhra Pradesh 2 121 Tirupathi Laddu Food stuff Andhra Pradesh 3 433 Bandar Laddu Food Stuff Andhra Pradesh 4 375 Arunachal Orange Agricultural Arunachal Pradesh 5 115 &118 Assam (Orthodox) Agricultural Assam 6 435 Assam Karbi Anglong Agricultural Assam Ginger 7 438 Tezpur Litchi Agricultural Assam 8 439 Joha Rice of Assam Agricultural Assam 9 558 Boka Chaul Agricultural Assam 10 609 Kaji Nemu Agricultural Assam 11 572 Chokuwa Rice of Assam Agricultural Assam 12 551 Bhagalpuri Zardalu Agricultural Bihar 13 553 Katarni Rice Agricultural Bihar 14 554 Magahi Paan Agricultural Bihar 15 552 Shahi Litchi of Bihar Agricultural Bihar 16 584 Silao Khaja Food Stuff Bihar 17 611 Jeeraphool Agricultural Chhattisgarh 18 618 Khola Chilli Agricultural Goa 19 185 Gir Kesar Mango Agricultural Gujarat 20 192 Bhalia Wheat Agricultural Gujarat 21 25 Kangra Tea Agricultural Himachal Pradesh 22 432 Himachali Kala Zeera Agricultural Himachal Pradesh 23 85 Monsooned Malabar Agricultural India Arabica Coffee (Karnataka & Kerala) 24 49 & 56 Malabar Pepper Agricultural India (Kerala, Karnataka & Tamilnadu) 25 385 Nagpur Orange Agricultural India (Maharashtra & Madhya Pradesh) 26 145 Basmati Agricultural India (Punjab / Haryana / Himachal Pradesh / Delhi / Uttarkhand / Uttar Pradesh / Jammu & Kashmir) 27 241 Banaganapalle Mangoes Agricultural India (Telangana & Andhra -

Registration Details of Geographical Indications
REGISTRATION DETAILS OF GEOGRAPHICAL INDICATIONS Goods S. Application Geographical Indications (As per Sec 2 (f) State No No. of GI Act 1999 ) FROM APRIL 2004 – MARCH 2005 Darjeeling Tea (word & 1 1 & 2 Agricultural West Bengal logo) 2 3 Aranmula Kannadi Handicraft Kerala 3 4 Pochampalli Ikat Handicraft Telangana FROM APRIL 2005 – MARCH 2006 4 5 Salem Fabric Handicraft Tamil Nadu 5 7 Chanderi Sarees Handicraft Madhya Pradesh 6 8 Solapur Chaddar Handicraft Maharashtra 7 9 Solapur Terry Towel Handicraft Maharashtra 8 10 Kotpad Handloom fabric Handicraft Odisha 9 11 Mysore Silk Handicraft Karnataka 10 12 Kota Doria Handicraft Rajasthan 11 13 & 18 Mysore Agarbathi Manufactured Karnataka 12 15 Kancheepuram Silk Handicraft Tamil Nadu 13 16 Bhavani Jamakkalam Handicraft Tamil Nadu 14 19 Kullu Shawl Handicraft Himachal Pradesh 15 20 Bidriware Handicraft Karnataka 16 21 Madurai Sungudi Handicraft Tamil Nadu 17 22 Orissa Ikat Handicraft Odisha 18 23 Channapatna Toys & Dolls Handicraft Karnataka 19 24 Mysore Rosewood Inlay Handicraft Karnataka 20 25 Kangra Tea Agricultural Himachal Pradesh 21 26 Coimbatore Wet Grinder Manufactured Tamil Nadu 22 28 Srikalahasthi Kalamkari Handicraft Andhra Pradesh 23 29 Mysore Sandalwood Oil Manufactured Karnataka 24 30 Mysore Sandal soap Manufactured Karnataka 25 31 Kasuti Embroidery Handicraft Karnataka Mysore Traditional 26 32 Handicraft Karnataka Paintings 27 33 Coorg Orange Agricultural Karnataka 1 FROM APRIL 2006 – MARCH 2007 28 34 Mysore Betel leaf Agricultural Karnataka 29 35 Nanjanagud Banana Agricultural -

A Study on Problems Faced by Paddy Cultivators in Salem District
INTERNATIONAL JOURNAL OF SCIENTIFIC & TECHNOLOGY RESEARCH VOLUME 9, ISSUE 03, MARCH 2020 ISSN 2277-8616 A Study On Problems Faced By Paddy Cultivators In Salem District Dr.M.Suguna, M.Jayanthi Abstract— Agriculture sector plays a vital role in Indian economy and it is the back bone of our country. India is endowed with land and water resources with conductive agro-climatic advantages for cultivation of paddy. Paddy is mainly grown in rain fed areas which receives heavy annual rainfall; it is primarily a kharif crop in India. Paddy is the important food crop in India; importance of paddy in agricultural crops cannot be ignored as it is the staple food for more than fifty percentage of population in the world. This study examines to identify the factors influencing paddy cultivation, to determine cultivating problems and marketing problems of paddy in the district of Salem in Tamil Nadu. Pre-scheduled interview questionnaire is prepared and circulated among the 84 respondents using convenient sampling. The results were obtained by application of statistical tools like percentage analysis and one sample t-test. Keywords—Agricultural sector, Cultivation problems, Indian economy, Marketing problems, Paddy production. ———————————————————— 1 INTRODUCTION Thalaivasal and Attur are the major concentration of paddy The Agricultural sector in India still contributes to the overall and high paddy concentrations of paddy and high paddy Attur, growth of the economy and provides livelihood and food Thalaivasal and Sankagiri. Many agricultural products security to a majority of the population. Monitoring the rice produced from Salem, a wide spread marketing throughout the market is a critical task considering more than half of the country. -
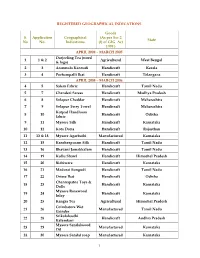
1 REGISTERED GEOGRAPHICAL INDICATIONS S. No Application No
REGISTERED GEOGRAPHICAL INDICATIONS Goods S. Application Geographical (As per Sec 2 State No No. Indications (f) of GIG Act 1999 ) APRIL 2004 – MARCH 2005 Darjeeling Tea (word 1 1 & 2 Agricultural West Bengal & logo) 2 3 Aranmula Kannadi Handicraft Kerala 3 4 Pochampalli Ikat Handicraft Telangana APRIL 2005 – MARCH 2006 4 5 Salem Fabric Handicraft Tamil Nadu 5 7 Chanderi Sarees Handicraft Madhya Pradesh 6 8 Solapur Chaddar Handicraft Maharashtra 7 9 Solapur Terry Towel Handicraft Maharashtra Kotpad Handloom 8 10 Handicraft Odisha fabric 9 11 Mysore Silk Handicraft Karnataka 10 12 Kota Doria Handicraft Rajasthan 11 13 & 18 Mysore Agarbathi Manufactured Karnataka 12 15 Kancheepuram Silk Handicraft Tamil Nadu 13 16 Bhavani Jamakkalam Handicraft Tamil Nadu 14 19 Kullu Shawl Handicraft Himachal Pradesh 15 20 Bidriware Handicraft Karnataka 16 21 Madurai Sungudi Handicraft Tamil Nadu 17 22 Orissa Ikat Handicraft Odisha Channapatna Toys & 18 23 Handicraft Karnataka Dolls Mysore Rosewood 19 24 Handicraft Karnataka Inlay 20 25 Kangra Tea Agricultural Himachal Pradesh Coimbatore Wet 21 26 Manufactured Tamil Nadu Grinder Srikalahasthi 22 28 Handicraft Andhra Pradesh Kalamkari Mysore Sandalwood 23 29 Manufactured Karnataka Oil 24 30 Mysore Sandal soap Manufactured Karnataka 1 25 31 Kasuti Embroidery Handicraft Karnataka Mysore Traditional 26 32 Handicraft Karnataka Paintings 27 33 Coorg Orange Agricultural Karnataka APRIL 2006 – MARCH 2007 28 34 Mysore Betel leaf Agricultural Karnataka 29 35 Nanjanagud Banana Agricultural Karnataka 30 37 Madhubani -

Government of India Ministry of Commerce & Industry Department for Promotion of Industry and Internal Trade
GOVERNMENT OF INDIA MINISTRY OF COMMERCE & INDUSTRY DEPARTMENT FOR PROMOTION OF INDUSTRY AND INTERNAL TRADE LOK SABHA UNSTARRED QUESTION NO. 3954. TO BE ANSWERED ON WEDNESDAY, THE 18TH MARCH, 2020. GRANTING GI-TAG 3954. DR. SUKANTA MAJUMDAR: DR. JAYANTA KUMAR ROY: SHRIMATI SANGEETA KUMARI SINGH DEO: SHRI RAJA AMARESHWARA NAIK: SHRI VINOD KUMAR SONKAR: SHRI BHOLA SINGH: Will the Minister of COMMERCE AND INDUSTRY be pleased to state: वाणि煍य एवं उद्योग मंत्री (a) whether the Government grants Geographical Indicators (GI)-tag to agricultural, natural or manufactured goods; (b) if so, the list of products with GI Logo issued/ to be issued, product-wise and State-wise; (c) whether the Government has ratified any international Intellectual Property (IP) treaty referred to in the International Intellectual Property Index (IIPI); (d) if so, whether the Government has launched the ‘Scheme for Intellectual Property Rights (IPR) Awareness – Creative India; Innovative India’; and (e) if so, the steps taken by the Government to improve the IPR regime in the country? ANSWER वाणि煍य एवं उद्योग मंत्री (श्री पीयूष गोयल) THE MINISTER OF COMMERCE & INDUSTRY (SHRI PIYUSH GOYAL) (a) & (b): Yes, sir, agricultural, natural or manufactured goods are registered as Geographical Indications (GI) by the Geographical Indications Registry as per the provisions of the Geographical Indications of Goods (Registration & Protection) Act, 1999. As on 10.03.2020, 362 Indian products have been registered as GIs. The list of GI products registered State-wise is enclosed as Annexure. (c): A private entity namely, Global Intellectual Property Centre of the U.S. -

FOOD PROCESSING TECHNOLOGY of RICE in ASSAM Dr.Sanjita Chetia Head, Dept.Of Home Science Digboi Mahila Mahavidyalaya Digboi
International Journal of Management, IT & Engineering Vol. 9 Issue 4, April 2019, ISSN: 2249-0558 Impact Factor: 7.119 Journal Homepage: http://www.ijmra.us, Email: [email protected] Double-Blind Peer Reviewed Refereed Open Access International Journal - Included in the International Serial Directories Indexed & Listed at: Ulrich's Periodicals Directory ©, U.S.A., Open J-Gate as well as in Cabell’s Directories of Publishing Opportunities, U.S.A FOOD PROCESSING TECHNOLOGY OF RICE IN ASSAM Dr.Sanjita Chetia Head, Dept.of Home science Digboi Mahila Mahavidyalaya Digboi -786171, Assam Abstract : Assam is traditionally a rice growing area. Rice plays a pivotal role in the socio-cultural life of the people of the state. The crop has enormous diversity in the region, which has resulted due to highly variable rice growing ecosystems. Besides, the region is inhabited by a large number of ethnic groups whose preference also varies from one another. All these factors are responsible for evolution of a large number of varieties in the region. Most of these have been in use from time immemorial with traditional method of preparation. Unknowingly people have selected many useful cultivars, which have commercial value in the present day world in which people prefer to have varieties of tastes. The science of food processing and preparation technique is based on the understanding of physical and chemical change that occur during processing. The knowledge can be used to combine food ingredient in diverse ways to prepare innumerable products with delicate flavours and colour which delight the senses.Food preparation technique is very much a part and parcel of theculture of the region. -

BY- ASHIRWAD SIR (Wifistudy UPSC)
2019 PIB 25.07.19 BY- ASHIRWAD SIR (wifistudy UPSC) CURRENT AFFAIRS PIB 25.07.19 BY-ASHIRWAD SIR Page 1 Cabinet approves creation of buffer stock of 40 LMT of sugar for a period of one year from 1st August 2019 to 31st July 2020 The Cabinet Committee on Economic Affairs (CCEA) chaired by Prime Minister Narendra Modi has approved the following proposals: 1. Creation of buffer stock of 40 lakh metric tonnes (LMT) of sugar for one year and to incur estimated maximum expenditure of Rs.1674 crores for this purpose. However, based on the market price and availability of sugar, this may be reviewed by the Deptt. of Food and Public Distribution any time for withdrawal / modification. 2. The reimbursement under the scheme would be met on quarterly basis to sugar mills which would be directly credited into farmers’ account on behalf of mills against cane price dues and subsequent balance, if any, would be credited to the mill’s account. Benefits: The decisions will lead to: 1. improvement in the liquidity of sugar mills; 2. reduction in sugar inventories; 3. stabilization in sugar prices by alleviating of price sentiments in domestic sugar market and thereby facilitate timely clearance of cane price dues of farmers; and 4. benefits for sugar mills in all sugarcane producing States, by way of clearing sugarcane price arrears of sugar mills Cabinet approves Determination of ‘Fair and Remunerative Price’ of sugarcane payable by sugar mills for 2019-20 sugar season The Cabinet Committee on Economic Affairs (CCEA) chaired by Prime Minister Narendra Modi has approved the proposal in respect of Determination of ‘Fair and Remunerative Price’ of sugarcane payable by sugar mills for 2019-20 sugar season. -

Agriculture Observer
Agriculture Observer www.agricultureobserver.com Volume :1 Issue :3 August 2020 Article No. :9 Trade Benefits of Agricultural Commodities Registered under GI Dhananjay N. Gawande1* and Dibendu Datta2 1Scientist, ICAR-National Research Centre for Grapes, Pune, Maharashtra. 2Principal Scientist, ICAR-Indian Institute of Pulses Research, Kanpur, Uttar Pradesh. Corresponding author*: [email protected] ABSTRACT Many agricultural produce pertaining to a specific geographical location have unique quality and taste. The trade interest of farmers producing such location specific premium quality produce can be protected through registration of these products under Geographical Indication (GI) of goods Act. If a product is registered under GI Act, only actual producers (as defined in the registry of the GI good) are eligible for doing commerce. GI Act prevents unauthorized producers from marketing the GI protected goods through legal protection and thus prevent the sale of spurious goods under GI banner. It provides competitive advantage to the producers and also offers consumers the genuine products. INTRODUCTION The marketing of agricultural goods are driven by consumer preferences. Basmati rice and Darjeeling tea fetches premium price for possessing special aroma and flavor. Quality of produce offers the producer the competitive advantage in market. GI Act was placed with the objective to protect the interest of real producers of quality goods originating from a specific geographic territory. Effective implementation of GI act at international level is important to benefit the local people associated with the production of GI goods through commercialization of the products. Genesis of GI in India Consequent upon India joining as a member state of the TRIPS Agreement (Trade Related Aspects of Intellectual Property Rights); a sui generis legislation for the protection of Geographical Indications was enacted in 1999. -

1St Indian Rice Congress
1st Indian Rice Congress - 2020 Rice Research and Development for Achieving Sustainable Development Goals Theme - I : Enhancing rice productivity and quality IRC/TM-1/PP-86 IDENTIFICATION OF GENOMIC REGIONS LINKED TO DORMANCY RELATED TRAITS USING BULK SEGREGANT ANALYSIS IN RICE (ORYZA SATIVA L.) D. B. K. V. Mani*, B. Krishna Veni, V. Roja and S. N. Umar Department of Genetics and Plant Breeding, Agricultural College, Bapatla-522101, Andhra Pradesh, India *Corresponding authors email: [email protected] A total of 119 F6:7 recombinant inbred lines of be associated with dormancy. Out of the 4 polymorphic a cross between BPT 2231 (non-dormant parent) and markers used in the present study, RM346 is notified MTU 1001 (dormant parent) was analyzed to identify as a dormancy linked marker from previous studies. the markers associated with dormancy. Parental The other 3 markers viz., RM22565, RM7051 and polymorphism survey with 188 SSR markers revealed RM10793 identified as dormancy linked markers in 10 polymorphic markers between the parents. The bulk the present study needs further validation on alternative segregant analysis results revealed that four markers set of population or a set of germplasm lines for their showed polymorphism between these bulks. The further utilization in the marker assisted breeding association of putative markers viz., RM346, programme. Based on germination percentage, RM22565, RM7051 and RM10793 identified based physiological parameters and genotyping studies, the on DNA pooling from selected segregant was analyzed lines SD 3, SD 12, SD 15 and SD 68 may be used in by Single Marker Analysis (SMA).The results of SMA future breeding programme as donor parents for seed revealed that RM22565 on chromosome 8 showed dormancy. -
Kisan Rath App for Assam
17TH ISSUE 17th IssueOctober | October 2020 2020 Kisan Rath App for Assam Kisan Rath App, customized for Assam to facilitate trade in fruits and vegetables was launched by the Hon’ble Chief Minister of Assam on 6th October 2020 at a function organized by the Assam Rural Infrastructure and Agricultural Services (ARIAS) Society, at Assam Administrative College, Khanapara, Guwahati. The KisanRath App was earlier launched by the Ministry of Agriculture and Farmers Welfare, Govt. of India in April 2020 in 8 languages to provide logistic support for farmers. The app was customized for Assam by the Assam Agribusiness and Rural Transformation Project (APART) and National Informatics Cell (NIC), Assam. APART has also built the capacity of the district Officials and project team by conducting online training 1 Assam Agribusiness and Rural Transformation Project 17th Issue | October 2020 sessions to all the districts. The pool of master trainers will support in disseminating information and also provide necessary guidance to the farmers, traders, FPOs, and others in downloading and installation of the KisanRath application. The App is aimed at providing better marketing linkage for Agri Horti produces like fruits, vegetables, spice crops etc. It will benefit the producer farmers, farmer producer organizations (FPOs), traders and service providers associated with Fruits and Vegetable marketing. As the app will directly link the producers with the traders of different markets of the state and outside, it is expected to minimize the involvement of middlemen in the marketing of Fruits and Vegetables and thereby provide better price to the producers. Farmers can offer their products for sale through this app by sharing the details viz., farmers name, place, contact number, product name, available quantity and offer price, etc, so that the traders dealing with those commodities may directly connect for procuring the items. -

Paddy in the World
IMPORTANT TRADITIONAL PADDY CULTIVARS OF NORTH EAST INDIA PADDY LOCATION CHARACTERISTICS PHOTO VARITIES A. Joha rice in the state are short to medium slender/ bold grain unlike Basmati Assam JOHA RICE rice which are long slender grains. ( Aromatic rice) B. This indigenous cultivars cook non sticky and tasty and gives higher return. C. The elongation ratio of joha rice 1.4 times. D. Aroma of Joha rice is high as Basmati E. There are a number of Joha cultivars like Kolajoha, Krishnajoha, Kunkunijoha, Ramphaljoha and Gobindbhog are important and widely grown cultivars in the state. Ketekijoha is the first HYV developed by Assam Agricultural University with high yield potential and aroma. F. It is grown in around 20,000ha producing around 30,000MT every year. Joha is sold @ Rs 40-75 in the retail markets. In 2007, the first consignment of 17MT of joha rice was exported to three European countries viz., Germany, U.K. and Switzerland where it was of high demand JOHA RICE A. Chokowa or soft rice: This is another class of rice used for instant CHAKOWA RICE preparation. Similar class of rice is not known in other parts of the world. Its (Soft and semi preparations are very popular in community feasts and festivals in Assam. It waxy rice) has amylase content of about 15%. B. Komal chaol (soft rice) are prepared from this class of rice by boiling paddy followed by drying and dehusking. C. This preparation can be preserved for quite long time and can be consumed instantly by soaking the rice in cold or hot water for brief period of time and then consumed with sugar or molasses, milk or curd even with salts , oils and pickles and vegetables.