Draft Genome Sequence of Pseudomonas Extremaustralis Strain USBA-GBX 515 Isolated from Superparamo Soil Samples in Colombian
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules As Potent Antibacterial and Antifungal Agents
Synthesis and Biological Evaluation of Trisindolyl-Cycloalkanes and Bis- Indolyl Naphthalene Small Molecules as Potent Antibacterial and Antifungal Agents Dissertation Zur Erlangung des akademischen Grades doctor rerum naturalium (Dr. rer. nat.) Vorgelegt der Naturwissenschaftlichen Fakultät I Institut für Pharmazie Fachbereich für Pharmazeutische Chemie der Martin-Luther-Universität Halle-Wittenberg von Kaveh Yasrebi Geboren am 09.14.1987 in Teheran/Iran (Islamische Republik) Gutachter: 1. Prof. Dr. Andreas Hilgeroth (Martin-Luther-Universität Halle-Wittenberg, Germany) 2. Prof. Dr. Sibel Süzen (Ankara Üniversitesi, Turkey) 3. Prof. Dr. Michael Lalk (Ernst-Moritz-Arndt-Universität Greifswald, Germany) Halle (Saale), den 21. Juli 2020 Selbstständigkeitserklärung Hiermit erkläre ich gemäß § 5 (2) b der Promotionsordnung der Naturwissenschaftlichen Fakultät I – Institut für Pharmazie der Martin-Luther-Universität Halle-Wittenberg, dass ich die vorliegende Arbeit selbstständig und ohne Benutzung anderer als der angegebenen Hilfsmittel und Quellen angefertigt habe. Alle Stellen, die wörtlich oder sinngemäß aus Veröffentlichungen entnommen sind, habe ich als solche kenntlich gemacht. Ich erkläre ferner, dass diese Arbeit in gleicher oder ähnlicher Form bisher keiner anderen Prüfbehörde zur Erlangung des Doktorgrades vorgelegt wurde. Halle (Saale), den 21. Juli 2020 Kaveh Yasrebi Acknowledgement This study was carried out from June 2015 to July 2017 in the Research Group of Drug Development and Analysis led by Prof. Dr. Andreas Hilgeroth at the Institute of Pharmacy, Martin-Luther-Universität Halle-Wittenberg. I would like to thank all the people for their participation who supported my work in this way and helped me obtain good results. First of all, I would like to express my gratitude to Prof. Dr. Andreas Hilgeroth for providing me with opportunity to carry out my Ph.D. -

Novel Antimicrobial Agents Inhibiting Lipid II Incorporation Into Peptidoglycan Essay MBB
27 -7-2019 Novel antimicrobial agents inhibiting lipid II incorporation into peptidoglycan Essay MBB Mark Nijland S3265978 Supervisor: Prof. Dr. Dirk-Jan Scheffers Molecular Microbiology University of Groningen Content Abstract..............................................................................................................................................2 1.0 Peptidoglycan biosynthesis of bacteria ........................................................................................3 2.0 Novel antimicrobial agents ...........................................................................................................4 2.1 Teixobactin ...............................................................................................................................4 2.2 tridecaptin A1............................................................................................................................7 2.3 Malacidins ................................................................................................................................8 2.4 Humimycins ..............................................................................................................................9 2.5 LysM ........................................................................................................................................ 10 3.0 Concluding remarks .................................................................................................................... 11 4.0 references ................................................................................................................................. -
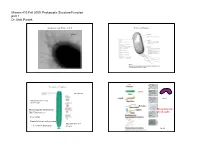
Parsek Micro410 Lecture
Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Organization of the Prokaryotic Cell Prokaryotic Structures fimbriae Size Range of Prokaryotes bacillus See Table 4.1 (rigid) vibrio Nanobacteria 0.05‐0.2 µm (0.14‐0.2 µm) (flexible) Thiomargarita namibiensis Mycoplasma are 700‐750 µm (Fig. 4.2) pleomorphic green alga Nanochlorum eukaryotum Mycoplasma 0.1‐ ~1-2 µm in diameter 0.3 µm Fig. 4.1 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Staining cells for Microscopic observation Cell Arrangements Motility- ~80% of prokaryotes are motile streptococcus Staining properties: Gram Stain staphylococcus Fig. 2.3 Gram Stain (1884) (Bacteria) Gram-negative mixed culture Gram-positive Fig. 2.3 and 2.4 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Functions of the cytoplasmic membrane The phospholipid bi‐layer Fig. 4.9 Fig. 4.4 What is the structure of bacterial phospholipids? Other components of the cytoplasmic membrane Figs. 4.5‐4.6 Microm 410 Fall 2009: Prokaryotic Structure/Function part 1 Dr. Matt Parsek Archaeal membranes can be a lipid monolayer Archaeal phospholipids have an ether linkage Fig. 4.7 Fig. 4.8 Importance of Cell Wall Schematic diagram cell wall • Provides rigidity to cell allowing cell to withstand the large osmotic/ionic Fig. 4.16 changes a bacterium may experience in its environment, and turgor pressure of cytoplasm (conc. of solutes in cytoplasm). Cell lysis • May have a role in shape determination. • Provides a barrier against certain toxic chemical and biological agents. • Site of action of some of the most commonly used antibiotics used to treat bacterial infections (penicillin family). -
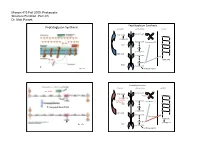
Parsek Lecture #3
Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Peptidoglycan Synthesis Peptidoglycan Synthesis cytoplasm cell membrane cell wall Bactoprenol-P Pi UDP-NAM M G pentapeptide G M Bactoprenol Bactoprenol-P-P P M UMP G P NAM G M pentapeptide M G UDP-NAG Bactoprenol G P NAM‐NAG P NAM-NAG UMP pentapeptide Fig. 6.7a Interbridge peptide Peptidoglycan Synthesis Cross-linking of Peptidoglycan Strands cytoplasm cell membrane cell wall autolysins Bactoprenol-P Pi UDP-NAM Bacitracin M G pentapeptide G D-cycloserine Bactoprenol M (Oxamycin) Bactoprenol-P-P P M UMP G P Transpeptidase (FtsI) NAM G M pentapeptide M G UDP-NAG Bactoprenol G Vancomycin P NAM‐NAG P NAM-NAG pentapeptide UMP Fig. 6.7b pentapeptide Interbridge peptide Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cross-linking of Peptidoglycan Strands Antibiotic Resistance autolysins • Inactivate antibiotic β-lactamase (penicillinase) Clavulanic acid β-lactams Augmentin and Trimentin (combination of clavulanic acid and transpeptidase amoxicillin or ampicillin respectively) penicillins and cephalosporins lysozyme • Change chemistry of target site • Limit access of the antibiotic to target site Fig. 6.5 Cell Shape Determination • Modifications made to Peptidoglycan: ‐ lysozyme: Protoplasts/spheroplasts ‐ autolysins Bacillus subtilis ‐ endopeptidase Heliobacter pylori • Protein(s) may play a major role ‐ MreB protein Caulobacter crescentus ‐ MreB has homology to actin, a component of the cytoskeleton of eukaryotes. Shape determining protein‐ crescentin Fig. 6.4 Microm 410 Fall 2009: Prokaryotic Structure/Function: Part 2/3 Dr. Matt Parsek Cell Wall Gram-positive Bacteria intermediate filaments in the bacteria Caulobacter crescentus glycerol similar predicted structures of crescentin and intermediate filaments Fig. -

APP201895 APP201895__Appli
APPLICATION FORM DETERMINATION Determine if an organism is a new organism under the Hazardous Substances and New Organisms Act 1996 Send by post to: Environmental Protection Authority, Private Bag 63002, Wellington 6140 OR email to: [email protected] Application number APP201895 Applicant Neil Pritchard Key contact NPN Ltd www.epa.govt.nz 2 Application to determine if an organism is a new organism Important This application form is used to determine if an organism is a new organism. If you need help to complete this form, please look at our website (www.epa.govt.nz) or email us at [email protected]. This application form will be made publicly available so any confidential information must be collated in a separate labelled appendix. The fee for this application can be found on our website at www.epa.govt.nz. This form was approved on 1 May 2012. May 2012 EPA0159 3 Application to determine if an organism is a new organism 1. Information about the new organism What is the name of the new organism? Briefly describe the biology of the organism. Is it a genetically modified organism? Pseudomonas monteilii Kingdom: Bacteria Phylum: Proteobacteria Class: Gamma Proteobacteria Order: Pseudomonadales Family: Pseudomonadaceae Genus: Pseudomonas Species: Pseudomonas monteilii Elomari et al., 1997 Binomial name: Pseudomonas monteilii Elomari et al., 1997. Pseudomonas monteilii is a Gram-negative, rod- shaped, motile bacterium isolated from human bronchial aspirate (Elomari et al 1997). They are incapable of liquefing gelatin. They grow at 10°C but not at 41°C, produce fluorescent pigments, catalase, and cytochrome oxidase, and possesse the arginine dihydrolase system. -

Genomic Analysis of Three Cheese-Borne Pseudomonas Lactis with Biofilm and Spoilage-Associated Behavior
microorganisms Article Genomic Analysis of Three Cheese-Borne Pseudomonas lactis with Biofilm and Spoilage-Associated Behavior Laura Quintieri 1 , Leonardo Caputo 1,* , Maria De Angelis 2 and Francesca Fanelli 1 1 Institute of Sciences of Food Production, National Research Council of Italy, Via G. Amendola 122/O, 70126 Bari, Italy; [email protected] (L.Q.); [email protected] (F.F.) 2 Department of Soil, Plant and Food Science, University of Bari Aldo Moro, Via Amendola 165/a, 70126 Bari, Italy; [email protected] * Correspondence: [email protected]; Tel.: +39-080-592-9323; Fax: +39-080-592-9374 Received: 10 July 2020; Accepted: 5 August 2020; Published: 8 August 2020 Abstract: Psychrotrophic pseudomonads cause spoilage of cold fresh cheeses and their shelf-life reduction. Three cheese-borne Pseudomonas sp., ITEM 17295, ITEM 17298, and ITEM 17299 strains, previously isolated from mozzarella cheese, revealed distinctive spoilage traits based on molecular determinants requiring further investigations. Genomic indexes (ANI, isDDH), MLST-based phylogeny of four housekeeping genes (16S rRNA, gyrB, rpoB and rpoD) and genome-based phylogeny reclassified them as Pseudomonas lactis. Each strain showed distinctive phenotypic traits at 15 and 30 ◦C: ITEM 17298 was the highest biofilm producer at both temperatures, whilst ITEM 17295 and ITEM 17299 showed the strongest proteolytic activity at 30 ◦C. A wider pattern of pigments was found for ITEM 17298, while ITEM 17295 colonies were not pigmented. Although the high genomic similarity, some relevant molecular differences supported this phenotypic diversity: ITEM 17295, producing low biofilm amount, missed the pel operon involved in EPS synthesis and the biofilm-related Toxin-Antitoxin systems (mqsR/mqsA, chpB/chpS); pvdS, required for the pyoverdine synthesis, was a truncated gene in ITEM 17295, harboring, instead, a second aprA involved in milk proteolysis. -

Pseudomonas Isolated from Spoiled Meat, Water, and Soil GORAN MOLIN and ANDERS TERNSTROM” Swedish Meat Research Institute, S-244 00 Kavlinge, Sweden
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, Apr. 1986, p. 257-274 Vol. 36, No. 2 0020-7713/86/020257-18$02.OO/O Copyright 0 1986, International Union of Microbiological Societies Phenotypically Based Taxonomy of Psychrotrophic Pseudomonas Isolated from Spoiled Meat, Water, and Soil GORAN MOLIN AND ANDERS TERNSTROM” Swedish Meat Research Institute, S-244 00 Kavlinge, Sweden The phenetic taxonomy of 305 strains of Pseudomonas and related organisms was numerically studied by using 215 features, including 156 assimilation tests. A total of 200 field strains were isolated from spoiling meat, and 50 strains were isolated from freshwater or soil. In addition, 55 reference strains (including 23 type strains and 4 clinical strains) were obtained. The strains clustered into 25 clusters at the 75% level when the Jaccard similarity coefficient was used. The 10 clusters that were considered significant were assigned to the Pseudomonas fragi complex (131 strains), Pseudomonas lundensis (40 strains), Pseudomonasfluorescens biovar 1 (27 strains), P.fluorescens biovar 2 (5 strains), P.fluorescens biovar 3 (6 strains), P.fluorescens biovar 4 (16 strains), Pseudomonas aureofaciens-Pseudomonas chlororaphis (3 strains), Pseudomonas aeruginosa (4 strains), Pseudomonas glathei (2 strains), and Pseudomonas mephitica (2 strains). The P. fragi complex was further divided into subclusters; the major subcluster (comprising 93 strains, including the type strain) was regarded as P. fragi sensu stricto. P. fluorescens and allied bacteria closely matched the descriptions given by Stanier et al. (J. Gen. Microbiol. 43:159-271, 1966). The characteristics for the 10 significant clusters are given. Also given are criteria which differentiate the P. fragi subclusters. The phylogenetic relationships among the meat-associated taxa were calculated, P.fluorescens biovars 2 and 3 were clearly separated from the remaining taxa. -

Download Article (PDF)
Biologia 66/2: 288—293, 2011 Section Cellular and Molecular Biology DOI: 10.2478/s11756-011-0021-6 The first investigation of the diversity of bacteria associated with Leptinotarsa decemlineata (Coleoptera: Chrysomelidae) Hacer Muratoglu, Zihni Demirbag &KazimSezen* Karadeniz Technical University, Faculty of Arts and Sciences, Department of Biology, 61080 Trabzon, Turkey; e-mail: [email protected] Abstract: Colorado potato beetle, Leptinotarsa decemlineata (Say), is a devastating pest of potatoes in North America and Europe. L. decemlineata has developed resistance to insecticides used for its control. In this study, in order to find a more effective potential biological control agent against L. decemlineata, we investigated its microbiota and tested their insecticidal effects. According to morphological, physiological and biochemical tests as well as 16S rDNA sequences, microbiota was identified as Leclercia adecarboxylata (Ld1), Acinetobacter sp. (Ld2), Acinetobacter sp. (Ld3), Pseudomonas putida (Ld4), Acinetobacter sp. (Ld5) and Acinetobacter haemolyticus (Ld6). The insecticidal activities of isolates at 1.8×109 bacteria/mL dose within five days were 100%, 100%, 35%, 100%, 47% and 100%, respectively, against the L. decemlineata larvae. The results indicate that Leclercia adecarboxylata (Ld1) and Pseudomonas putida (Ld4) isolates may be valuable potential biological control agents for biological control of L. decemlineata. Key words: Leptinotarsa decemlineata; 16S rDNA; microbiota; insecticidal activity; microbial control. Abbreviations: ANOVA, one-way analysis of variance; LSD, least significant difference; PBS, phosphate buffer solution. Introduction used because of marketing concerns and limited num- ber of transgenic varieties available. Also, recombinant Potato is an important crop with ∼4.3 million tons defence molecules in plants may affect parasitoids or of production on 192,000 hectares of growing area predators indirectly (Bouchard et al. -

Comparative Analysis of the Core Proteomes Among The
Diversity 2020, 12, 289 1 of 25 Article Comparative Analysis of the Core Proteomes among the Pseudomonas Major Evolutionary Groups Reveals Species‐Specific Adaptations for Pseudomonas aeruginosa and Pseudomonas chlororaphis Marios Nikolaidis 1, Dimitris Mossialos 2, Stephen G. Oliver 3 and Grigorios D. Amoutzias 1,* 1 Bioinformatics Laboratory, Department of Biochemistry and Biotechnology, University of Thessaly, 41500 Larissa, Greece; [email protected] 2 Microbial Biotechnology‐Molecular Bacteriology‐Virology Laboratory, Department of Biochemistry and Biotechnology, University of Thessaly, 41500 Larissa, Greece; [email protected] 3 Cambridge Systems Biology Centre & Department of Biochemistry, University of Cambridge, Cambridge CB2 1GA, UK; [email protected] * Correspondence: [email protected]; Tel.: +30‐2410‐565289; Fax: +30‐2410‐565290 Received: 22 June 2020; Accepted: 22 July 2020; Published: 24 July 2020 Abstract: The Pseudomonas genus includes many species living in diverse environments and hosts. It is important to understand which are the major evolutionary groups and what are the genomic/proteomic components they have in common or are unique. Towards this goal, we analyzed 494 complete Pseudomonas proteomes and identified 297 core‐orthologues. The subsequent phylogenomic analysis revealed two well‐defined species (Pseudomonas aeruginosa and Pseudomonas chlororaphis) and four wider phylogenetic groups (Pseudomonas fluorescens, Pseudomonas stutzeri, Pseudomonas syringae, Pseudomonas putida) with a sufficient number of proteomes. As expected, the genus‐level core proteome was highly enriched for proteins involved in metabolism, translation, and transcription. In addition, between 39–70% of the core proteins in each group had a significant presence in each of all the other groups. Group‐specific core proteins were also identified, with P. -

Università Degli Studi Di Padova Dipartimento Di Biomedicina Comparata Ed Alimentazione
UNIVERSITÀ DEGLI STUDI DI PADOVA DIPARTIMENTO DI BIOMEDICINA COMPARATA ED ALIMENTAZIONE SCUOLA DI DOTTORATO IN SCIENZE VETERINARIE Curriculum Unico Ciclo XXVIII PhD Thesis INTO THE BLUE: Spoilage phenotypes of Pseudomonas fluorescens in food matrices Director of the School: Illustrious Professor Gianfranco Gabai Department of Comparative Biomedicine and Food Science Supervisor: Dr Barbara Cardazzo Department of Comparative Biomedicine and Food Science PhD Student: Andreani Nadia Andrea 1061930 Academic year 2015 To my family of origin and my family that is to be To my beloved uncle Piero Science needs freedom, and freedom presupposes responsibility… (Professor Gerhard Gottschalk, Göttingen, 30th September 2015, ProkaGENOMICS Conference) Table of Contents Table of Contents Table of Contents ..................................................................................................................... VII List of Tables............................................................................................................................. XI List of Illustrations ................................................................................................................ XIII ABSTRACT .............................................................................................................................. XV ESPOSIZIONE RIASSUNTIVA ............................................................................................ XVII ACKNOWLEDGEMENTS .................................................................................................... -

16S Rdna Analysis for Characterization of Pesudomonas Sp
16S rDNA analysis for characterization of Pseudomonas sp. strain MF30 isolated from Rumex acetocella roots in northern Sweden (Received: 15.09 .2006; Accepted: 28.09.2006) Idress H. Attitalla*;Ali A. Bataw**; Jolanta J. Borowicz***and Sture Brishammar*** *Omar Al-Mukhtar University, Faculty of Science, Botany Department, Box 919, El-Beida, Libya **Omar Al-Mukhtar University, Faculty of Science, Zoology Department, Box 919, El-Beida, Libya ***Maselaboratoratorerna AB, Box 148, Uppsala, Sweden Corresponding author1. [email protected] ABSTRACT A bacterial strain obtained from the northern part of Sweden previously classified as Pseudomonas veronii based on biochemical and physioloical tests. In this study, phylogenetic tree was constructed using a nearly complete sequence within the 16S rDNA gene. The strain of Pseudomonas sp. subdivided into two rather distinctly related groups, neither of which is very close to the group within the Pseudomonas fluorescens cluster. Although the phylogenetic analysis is not conclusive, it is consistent with other observations, especially the capacities of the this strain as a biocontrol agent. Taken all together, the results suggest that the MF30 strain should be classifed as another Pseudomonas species, either Pseudomonas antarctica or P. meridina. Key words: Phylogenetic analysis, 16S rDNA gene, Pseudomonas species, P. veronii, P. antarctica, P. meridina. INTRODUCTION received attention even though they possess abilities to influence plant growth and acteria belonging to the fluorescent development through different mechanisms pseudomonads, known for the (Weller, 1988; O’Sullivan and O’Gara, 1992). B diversity of their metabolites They are now recognised as being antagonistic (Leisinger and Margaff, 1979), contain to several opportunistic soil-borne fungi species that are recognized as human and as (Weller, 1988; Keel et al., 1996) and to seed- animal pathogens (Nakazawa and Abe, 1996) borne fungi (Hökeberg et al., 1997). -

Pseudomonas Monteilii Sp. Nov., Isolated from Clinical Specimens
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, July 1997, p. 846-852 Vol. 47, No. 3 0020-7713/97/$04.00+0 Copyright 0 1997, International Union of Microbiological Societies Pseudomonas monteilii sp. nov., Isolated from Clinical Specimens MALIKA ELOMARI, LOIC COROLER, SOPHIE VERHILLE, DANIEL IZARD, AND HENRI LECLERC" Laboratoire de Bacteriologie Hygiine, Faculti de Midecine Henri Warembourg, 59045 Lille Cedex, France We propose the name Pseudomonas monteilii for a new species of gram-negative, rod-shaped, motile bacteria that were nonhemolytic on blood agar and were isolated from clinical sources. The 10 strains of P. monteilii were incapable of liquefing gelatin. They grew at 10°C but not at 41"C,produced fluorescent pigments, catalase, and cytochrome oxidase, and possessed the arginine dihydrolase system. They were capable of respiratory but not fermentative metabolism. They did not hydrolyze esculin or starch and were able to use benzylamine, a- aminobutyrate, D-ribose, L-arabinose,butyrate, valerate, isovalerate, isobutyrate, inositol, phenylacetate, D-ala- nine, and amylamine. They possessed L-phenylalanine arylamidase, L-lysine arylamidase, L-alanine arylamidase, y-glutamyl-transferase,glycyl-phenylalanine arylamidase, L-tryptophan arylamidase, glycyl-L-alaninearylami- dase, esterase C,, esterase C,, esterase C,, esterase C,, esterase Cl0,and esterase CIS.DNA relatedness studies revealed that P. monteilii strains formed a homogeneous DNA hybridization group. A total of 57 strains rep- resenting previously described or partially characterized taxa belonging to the genus Pseudomoms were 6 to 54% related to P. monteilii. The highest hybridization values were obtained with strains belonging to or related to Pseu- domonasputida biovar A. The average G+C content of the DNA was 60.5 0.5 mol%for four of the P.