Development of Intranasal Bacterial Therapeutics to Mitigate the Bovine Respiratory Pathogen Mannheimia Haemolytica
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ecology and Epidemiology of Campylobacter Jejuni in Broiler Chickens
Ecology and Epidemiology of Campylobacter jejuni in Broiler Chickens A DISSERTATION SUBMITTED TO THE FACULTY OF THE UNIVERSITY OF MINNESOTA BY Hae Jin Hwang IN PARTIALL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Dr. Randall Singer, Dr. George Maldonado June 2019 © Hae Jin Hwang, 2019 Acknowledgements I would like to sincerely thank my advisor, Dr. Randall Singer, for his intellectual guidance and support, great patience, and mentorship, which made this dissertation possible. I would also like to thank Dr. George Maldonado for his continuous encouragement and support. I would further like to thank my thesis committee, Dr. Richard Isaacson and Dr. Timothy Church, for their guidance throughout my doctoral training. I thank all my friends and colleagues I met over the course of my studies. I am especially indebted to my friends, Dr. Kristy Lee, Dr. Irene Bueno Padilla, Dr. Elise Lamont, Madhumathi Thiruvengadam, Dr. Kaushi Kanankege and Dr. Sylvia Wanzala, for their support and friendship. Heartfelt gratitude goes to my family, for always believing in me, encouraging me and helping me get through the difficult and stressful times during my studies. Lastly, I thank Sven and Bami for being the best writing companions I could ever ask for. i Abstract Campylobacteriosis, predominantly caused by Campylobacter jejuni, is a common, yet serious foodborne illness. With consumption and handling of poultry products as the most important risk factor of campylobacteriosis, reducing Campylobacter contamination in poultry products is considered the best public health intervention to reduce the burden and costs associated with campylobacteriosis. To this end, there is a need to improve our understanding of epidemiology and ecology of Campylobacter jejuni in poultry. -

A Taxonomic Note on the Genus Lactobacillus
Taxonomic Description template 1 A taxonomic note on the genus Lactobacillus: 2 Description of 23 novel genera, emended description 3 of the genus Lactobacillus Beijerinck 1901, and union 4 of Lactobacillaceae and Leuconostocaceae 5 Jinshui Zheng1, $, Stijn Wittouck2, $, Elisa Salvetti3, $, Charles M.A.P. Franz4, Hugh M.B. Harris5, Paola 6 Mattarelli6, Paul W. O’Toole5, Bruno Pot7, Peter Vandamme8, Jens Walter9, 10, Koichi Watanabe11, 12, 7 Sander Wuyts2, Giovanna E. Felis3, #*, Michael G. Gänzle9, 13#*, Sarah Lebeer2 # 8 '© [Jinshui Zheng, Stijn Wittouck, Elisa Salvetti, Charles M.A.P. Franz, Hugh M.B. Harris, Paola 9 Mattarelli, Paul W. O’Toole, Bruno Pot, Peter Vandamme, Jens Walter, Koichi Watanabe, Sander 10 Wuyts, Giovanna E. Felis, Michael G. Gänzle, Sarah Lebeer]. 11 The definitive peer reviewed, edited version of this article is published in International Journal of 12 Systematic and Evolutionary Microbiology, https://doi.org/10.1099/ijsem.0.004107 13 1Huazhong Agricultural University, State Key Laboratory of Agricultural Microbiology, Hubei Key 14 Laboratory of Agricultural Bioinformatics, Wuhan, Hubei, P.R. China. 15 2Research Group Environmental Ecology and Applied Microbiology, Department of Bioscience 16 Engineering, University of Antwerp, Antwerp, Belgium 17 3 Dept. of Biotechnology, University of Verona, Verona, Italy 18 4 Max Rubner‐Institut, Department of Microbiology and Biotechnology, Kiel, Germany 19 5 School of Microbiology & APC Microbiome Ireland, University College Cork, Co. Cork, Ireland 20 6 University of Bologna, Dept. of Agricultural and Food Sciences, Bologna, Italy 21 7 Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Vrije Universiteit 22 Brussel, Brussels, Belgium 23 8 Laboratory of Microbiology, Department of Biochemistry and Microbiology, Ghent University, Ghent, 24 Belgium 25 9 Department of Agricultural, Food & Nutritional Science, University of Alberta, Edmonton, Canada 26 10 Department of Biological Sciences, University of Alberta, Edmonton, Canada 27 11 National Taiwan University, Dept. -

The Mysterious Orphans of Mycoplasmataceae
The mysterious orphans of Mycoplasmataceae Tatiana V. Tatarinova1,2*, Inna Lysnyansky3, Yuri V. Nikolsky4,5,6, and Alexander Bolshoy7* 1 Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, 90027, California, USA 2 Spatial Science Institute, University of Southern California, Los Angeles, 90089, California, USA 3 Mycoplasma Unit, Division of Avian and Aquatic Diseases, Kimron Veterinary Institute, POB 12, Beit Dagan, 50250, Israel 4 School of Systems Biology, George Mason University, 10900 University Blvd, MSN 5B3, Manassas, VA 20110, USA 5 Biomedical Cluster, Skolkovo Foundation, 4 Lugovaya str., Skolkovo Innovation Centre, Mozhajskij region, Moscow, 143026, Russian Federation 6 Vavilov Institute of General Genetics, Moscow, Russian Federation 7 Department of Evolutionary and Environmental Biology and Institute of Evolution, University of Haifa, Israel 1,2 [email protected] 3 [email protected] 4-6 [email protected] 7 [email protected] 1 Abstract Background: The length of a protein sequence is largely determined by its function, i.e. each functional group is associated with an optimal size. However, comparative genomics revealed that proteins’ length may be affected by additional factors. In 2002 it was shown that in bacterium Escherichia coli and the archaeon Archaeoglobus fulgidus, protein sequences with no homologs are, on average, shorter than those with homologs [1]. Most experts now agree that the length distributions are distinctly different between protein sequences with and without homologs in bacterial and archaeal genomes. In this study, we examine this postulate by a comprehensive analysis of all annotated prokaryotic genomes and focusing on certain exceptions. -

1St Year : Immunity
Presented by-Suman Bampal Lecturer pharmacy Govt polytechnic, pithuwala Dehradun Introduction Immunology is the science which deals with immunity or resistance to body infection . The lack of ability to resist infection is called susceptibility Preparations use to produce immunity are called immunological preparations Factors affecting immunity 1. Phagocytosis : ingestion of microorganisms by certain cells (Phagocytes ) of the body whereby they are rendered harmless . It is caused by - W.B.C (leucocytes), Cells of R.E.S 2. Antibody production : these are highly specific in nature and attack microorganism or toxins . Antibodies are proteins mainly globulins produced in lymph nodes by the cells of R.E.S Nature of antibodies depends upon the manner in which microorganism produce their harmful effect Bacteria producing exotoxins - antitoxins Bacteria producing endotoxins – these antibodies are named according to their mode of action Antigen-Antibody Reaction Antigen antibody nature of reaction Exotoxin Antitoxin Neutrilization Bacterial cells Agglutinin Agglutination Endotoxin precipitin precipitation of toxin Bacterial cells Bacteriolysin * Lysis of cells . Opsonins Makes pathogens more susceptible to phagocytosis Bacteria + Specific Bacteriolysin – no lysis Bacteria + Complement – no lysis Bacteria + Specific Bacteriolysin + Complement – lysis of the bacteria. immunity 1.Natural immunity 2. Acquired immunity (God gifted) (acquired due to antibodies production) a .Species active passive b . Race (slowly produced but long lasting) (quickly prod. c. individual but short lived) d. Age Naturally acquired Naturally acquired (after infection) ( from mother through placenta) Artificially acquired Artificially acquired (due to admin. of vaccines or antigens) (by admin. of serum) 1.Natural Immunity Species – e.g. Tuberclosis is very fetal to guineapig but not fetal to man. -

Production of Antibodies for Use in a Biosensor- Based Assay for Listeria Monocytogenes
Production of antibodies for use in a biosensor- based assay for Listeria monocytogenes A thesis submitted for the degree of Ph.D. By Paul Leonard B.Sc. (Hons), August 2003. Based on research carried out at School of Biotechnology, Dublin City University, D ublin 9, Ireland, Under the supervision of Professor Richard O’Kennedy. This thesis is dedicated to my parents for all their encouragement and support over the last number of years. “I am not discouraged, because every wrong attempt discarded is another step forward” -Thomas Edison. Declaration I hereby certify that this material, which 1 now submit for assessment on the programme of study leading to the award of Doctor of Philosophy, is entirely my own work, and has not been taken from the work of others, save and to the extent that such work is cited and acknowledged within the text. Signed Date: Acknowledgements Sincere thanks to Prof. Richard O'Kennedy for his constant support and guidance over the past few years, in particular for sharing his wealth of experience and knowledge throughout my studies. Thanks to all the lab group, past and present and all my friends at DC’U for their companionship and some unforgettable (and more often than not better forgotten) nights out! Special thanks goes to Steve, a great scientist and friend, for his support, knowledge and overall enthusiasm over the last few years. To Monty, Macker, Ryaner and the rest of the “Finian's lads” for their continual ‘good humoured harassment’ and alcohol-based support! Cheers! I would like to thank all my family for their unequivocal support from start to finish, I owe you so much! Finally, 1 would like to reserve a very special thanks to Nerea, my source of inspiration, for her patience, companionship and constant love and support. -

Correlation Between the Oral Microbiome and Brain Resting State Connectivity in Smokers
bioRxiv preprint doi: https://doi.org/10.1101/444612; this version posted October 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Correlation between the oral microbiome and brain resting state connectivity in smokers Dongdong Lin1, Kent Hutchison2, Salvador Portillo3, Victor Vegara1, Jarod Ellingson2, Jingyu Liu1,3, Amanda Carroll-Portillo3,* ,Vince D. Calhoun1,3,* 1The Mind Research Network, Albuquerque, New Mexico, 87106 2University of Colorado Boulder, Boulder, CO 3University of New Mexico, Department of Electrical and Computer Engineering, Albuquerque, New Mexico, 87106 * authors contributed equally to the work. Abstract Recent studies have shown a critical role for the gastrointestinal microbiome in brain and behavior via a complex gut–microbiome–brain axis, however, the influence of the oral microbiome in neurological processes is much less studied, especially in response to the stimuli in the oral microenvironment such as smoking. Additionally, given the complex structural and functional networks in brain system, our knowledge about the relationship between microbiome and brain functions on specific brain circuits is still very limited. In this pilot work, we leverage next generation microbial sequencing with functional MRI techniques to enable the delineation of microbiome-brain network links as well as their relations to cigarette smoking. Thirty smokers and 30 age- and sex- matched non-smokers were recruited for measuring both microbial community and brain functional networks. Statistical analyses were performed to demonstrate the influence of smoking on: the taxonomy and abundance of the constituents within the oral microbial community, brain functional network connectivity, and associations between microbial shifts and the brain signaling network. -

UNIT 3 IMMUNITY Structure
Microbiology-II UNIT 3 IMMUNITY Structure 3.0 Objectives 3.1 Introduction 3.2 Definitions 3.3 What is Immunity? 3.4 The Three Lines of Defense in the Body 3.5 Inflammation 3.6 Types of Immunity 3.6.1 Innate Immunity 3.6.2 Factors Influencing Innate Immunity in an Individual 3.6.3 Acquired Immunity 3.6.4 Active Acquired Immunity 3.6.5 Passive Acquired Immunity 3.6.6 Differences between Active and Passive Immunity 3.7 The Immune System 3.8 Antigens and Antibodies 3.9 Allergy/Hypersensitivity/Anaphylaxis 3.10 Practical Application of Immunology 3.10.1 Immunizing Agents 3.10.2 Vaccines and Vaccinations 3.10.3 Immunoglobulins 3.10.4 The Immune Responses 3.11 Let Us Sum Up 3.12 Key Words 3.13 Answers to Check Your Progress 3.0 OBJECTIVES After going through this unit, you should be able to: l define Immunity; l differentiate between the older and modern concept of immunity; l determine the three lines of defense in the body; l enumerate the different types of immunity; l differentiate between active and passive immunity; l explain the innate and adaptive immune system; l distinguish between antigens and antibodies; l state the phenomenon of allergy; l describe the various antigen-antibody reactions; l discuss the various immunizing agents; and l define primary and secondary response. 3.1 INTRODUCTION The word ‘Immunity’ comes from a Latin term ‘immunis’ meaning free or exempt. It has been recognised from very early times that those who suffer from infectious diseases such 38 as diphtheria, whooping cough, mumps, measles, small pox etc. -
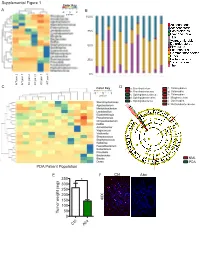
Mental Figure 1 Color Key a -2 0 2 B Z-Score 100%
Supplemental Figure 1 Color Key A -2 0 2 B z-score 100% 75% 50% 25% 0% KC pan 1 WT pan 3 WT KC pan 3 WT pan 2 WT pan 1 WT KC pan 2 C Color Key D a: Brevibacterium f: Chlamydiales b: Brevibacteriaceae g: Chlamydiia -3 0 3 z-score c: Sphingobacteriaceae h: Chlamydiae d: Sphingobacteriales i: Mogibacterium e: Sphingobacteriia j: Oscillospira k: Methylobacteriaceae NML Control Microb.-entrained MΦ PDA PDA Patient Population Control Microb.-entrained MΦ + Myd88i E F Ctrl Abx 350 * 300 250 200 150 40X 100 Tumor weight (mg) 50 0 x Ctrl Ab Supplemental Figure 2 A KC WT B ** * Actinobacteria * ** Bacteroidetes Cyanobacteria Deferribacteres * Firmicutes Proteobacteria % Relative abundance TM7 Others Time(wks) 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 Alpha Diversity Measure C E 60 KC WT 40 20 B. pseudolongum B. animalis 60 5 KC WT 0 B. adolescentis 40% Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 20 Age (weeks) B. pseudolongum B. animalis 5 0 B. adolescentis % Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 F Age (weeks) Week 3 Week 9 Week 13 p=0.678 p=0.02 p=0.385 Time(wks) 3 9 24 20 13 16 D 28 32 36 Week 13 KC WT Firmicutes; Ruminococcus Firmicutes; Dehalobacterium Alpha Diversity Measure Firmicutes; Oscillospira Bacteroidates; Odoribacter Axis.2 [12.7%] Actinobacteria; Bifidobacterium Axis.2 [23.8%] Axis.2 [24.7%] Week 16 Bacteroidetes; Bacteroidales Axis.1 [80.8%] Axis.1 [65.4%] Axis.1 [49.6%] Actinobacteria; Bifidobacterium Week 16 Week 20 Week 24 Week 20 p=0.339 p=0.036 p=0.021 Firmicutes; Dehalobacterium -

Exploring the Cockatiel (Nymphicus Hollandicus) Fecal Microbiome, Bacterial Inhabitants of a Worldwide Pet
Exploring the cockatiel (Nymphicus hollandicus) fecal microbiome, bacterial inhabitants of a worldwide pet Luis David Alcaraz1, Apolinar M. Hernández2 and Mariana Peimbert2 1 Laboratorio Nacional de Ciencias de la Sostenibilidad, Instituto de Ecología, Universidad Nacional Autonóma de México, Mexico City, Mexico 2 Departamento de Ciencias Naturales, Unidad Cuajimalpa, Universidad Autónoma Metropolitana, Mexico City, Mexico ABSTRACT Background. Cockatiels (Nymphicus hollandicus) were originally endemic to Australia; now they are popular pets with a global distribution. It is now possible to conduct detailed molecular studies on cultivable and uncultivable bacteria that are part of the intestinal microbiome of healthy animals. These studies show that bacteria are an essential part of the metabolic capacity of animals. There are few studies on bird microbiomes and, to the best of our knowledge, this is the first report on the cockatiel microbiome. Methods. In this paper, we analyzed the gut microbiome from fecal samples of three healthy adult cockatiels by massive sequencing of the 16S rRNA gene. Additionally, we compared the cockatiel fecal microbiomes with those of other bird species, including poultry and wild birds. Results. The vast majority of the bacteria found in cockatiels were Firmicutes, while Proteobacteria and Bacteroidetes were poorly represented. A total of 19,280 different OTUs were detected, of which 8,072 belonged to the Erysipelotrichaceae family. Discussion. It is relevant to study cockatiel the microbiomes of cockatiels owing to their wide geographic distribution and close human contact. This study serves as a reference for cockatiel bacterial diversity. Despite the large OTU numbers, the diversity is not even Submitted 14 July 2016 Accepted 28 November 2016 and is dominated by Firmicutes of the Erysipelotrichaceae family. -

MULTI-FUNCTIONAL EFFECTOR RESPONSES ELICITED from Igm MEMORY STEM CELLS
MULTI-FUNCTIONAL EFFECTOR RESPONSES ELICITED FROM IgM MEMORY STEM CELLS Item Type Dissertation Authors Kenderes, Kevin Rights Attribution-NonCommercial-NoDerivatives 4.0 International Download date 29/09/2021 18:50:57 Item License http://creativecommons.org/licenses/by-nc-nd/4.0/ Link to Item http://hdl.handle.net/20.500.12648/2017 MULTI-FUNCTIONAL EFFECTOR RESPONSES ELICITED FROM IgM MEMORY STEM CELLS by Kevin Kenderes A Dissertation in Microbiology and Immunology Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Graduate Studies of State University of New York, Upstate Medical University. Approved ___________________________ (Sponsor’s signature) Date_______________________ Table of Contents List of Figures iv List of Tables vi Abbreviations vii Acknowledgements x Abstract xi Chapter I: Introduction 1 History of Humoral Immunity 2 Development of Naive B cells 4 The Primary B cell Response 7 T cell dependent B cell responses 7 T cell independent B cell responses 10 Secondary B cell Responses 12 Ehrlichia muris 15 Immune Responses to E. muris 17 T-bet-Expressing B Cells 19 Summary 21 Chapter II: Materials and Methods 22 Chapter III: CD11c+ T-bet+ IgM memory Cells Exhibit Stem Cell-like 27 Properties Abstract 28 Introduction 29 Results 33 ii Discussion 76 Chapter IV: Summary 84 Model 85 Future directions 90 Summary 95 Appendix I: Tetanus Toxin binds to bystander B cells following immunization 97 Abstract 98 Introduction 99 Materials and Methods 102 Results 105 Discussion 124 Future Directions 128 References 130 iii List of Figures Figure 3.1. Labeling Aicda-expressing CD11c+ T-bet+ IgM memory cells in vivo 34 Figure 3.S1. -

Prevalence of Ureaplasma Urealyticum, Mycoplasma Hominis and Chlamydia Trachomatis in Patients with Uncomplicated Recurrent Urin
Nephrology and Renal Diseases Research Article ISSN: 2399-908X Prevalence of Ureaplasma urealyticum, Mycoplasma hominis and Chlamydia trachomatis in patients with uncomplicated recurrent urinary tract infections Jadranka Vlasic-Matas1*, Hrvoje Raos2, Marijana Vuckovic2, Stjepan Radic2 and Vesna Capkun3 1Polyclinic Nephrology Department, Split, Croatia 2School of Medicine, University of Split, Split, Croatia 3Department of Nuclear Medicine, Split University Hospital Center, Split, Croatia Abstract Aim: To assess the prevalence of Ureaplasma urealyticum, Mycoplasma hominis and Chlamydia trachomatis in patients with chronic urinary tract infections (UTIs) and its correlation with leukocyturia and symptoms. Methods: The study included 220 patients (130 women and 90 men) presenting with chronic voiding symptoms and sterile leukocyturia. Urine, urethral swabs and cervical swabs (for women patients) were taken to determine the presence of these pathogens. Patients were treated by tetracycline and followed up three and six months after initial therapy. Results: In 186 (85%) out of 220 patients, U. urealyticum was found, while C. trachomatis was present in 34 patients (15%). In majority of female patients (112 out of 130; 86%) U. urealyticum was found. In addition to ureaplasma, in eight patients M. hominis was found. C. trachomatis was identified in 18 female patients (14%). In 74 out of 90 (82%) male patients U. urealyticum was detected while in six of them M. hominis was also found. C. trachomatis was identified in 16 male patients (18%). U. urealyticum was significantly related to leukocyturia, as opposed to C. trachomatis (p<0,001). Women had more frequent symptomatology (p = 0,015) and higer leukocyturia (p<0.001). Conclusion: Leukocyturia is more common find in U. -

Amarnath VS Pisipati Doctor of Philosophy
SINGLE, ULTRA-HIGH DOSE AMINOGLYCOSIDE THERAPY IN A RAT MODEL OF E. COLI INDUCED SEPTIC SHOCK By Amarnath V. S. Pisipati A Thesis Submitted to the Faculty of Graduate Studies of The University of Manitoba In Partial Fulfillment of the Requirements of the Degree of Doctor of Philosophy Department of Medical Microbiology University of Manitoba, Winnipeg, Manitoba, Canada Copyright © 2015 by Amarnath V. S. Pisipati II ABSTRACT Bacterial infections are a major cause of morbidity and mortality in both the community and nosocomial settings, particularly among the elderly and chronically ill. Sepsis is the body’s response to antigens and toxins released by the invasive pathogenic organisms that cause infection. When infection is not effectively controlled, sepsis may develop and progress to severe sepsis and septic shock. Early diagnosis and treatment is pivotal for survival in severe sepsis and particularly, septic shock. Our research focuses on developing a novel treatment strategy for septic shock by using single, ultra-high doses of aminoglycosides. In this project, the effect of a single, ultra-high dose of gentamicin in clearing bacteria from the blood and reducing the bacterial burden in vital organs was evaluated in a rat model of E. coli (Bort strain) induced peritonitis with severe sepsis/septic shock. Serum cytokine levels and serum lactate levels were serially measured. Further, the potential adverse effects of ultra-high dosing of aminoglycoside antibiotics in a short-term (9 h) invasive study and long- term (180 days) non-invasive study were assessed. Neuromuscular paralyses due to ultra-high doses of aminoglycosides were assessed. In addition, renal injury markers such as serum creatinine and urinary Neutrophil Gelatinase Associated Lipocalin (NGAL) were assayed.