Brook Milligan NMSU.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Project Rapid-Field Identification of Dalbergia Woods and Rosewood Oil by NIRS Technology –NIRS ID
Project Rapid-Field Identification of Dalbergia Woods and Rosewood Oil by NIRS Technology –NIRS ID. The project has been financed by the CITES Secretariat with funds from the European Union Consulting objectives: TO SELECT INTERNATIONAL OR NATIONAL XYLARIUM OR WOOD COLLECTIONS REGISTERED AT THE INTERNATIONAL ASSOCIATION OF WOOD ANATOMISTS – IAWA THAT HAVE A SIGNIFICANT NUMBER OF SPECIES AND SPECIMENS OF THE GENUS DALBERGIA TO BE ANALYZED BY NIRS TECHNOLOGY. Consultant: VERA TERESINHA RAUBER CORADIN Dra English translation: ADRIANA COSTA Dra Affiliations: - Forest Products Laboratory, Brazilian Forest Service (LPF-SFB) - Laboratory of Automation, Chemometrics and Environmental Chemistry, University of Brasília (AQQUA – UnB) - Forest Technology and Geoprocessing Foundation - FUNTEC-DF MAY, 2020 Brasília – Brazil 1 Project number: S1-32QTL-000018 Host Country: Brazilian Government Executive agency: Forest Technology and Geoprocessing Foundation - FUNTEC Project coordinator: Dra. Tereza C. M. Pastore Project start: September 2019 Project duration: 24 months 2 TABLE OF CONTENTS 1. INTRODUCTION 05 2. THE SPECIES OF THE GENUS DALBERGIA 05 3. MATERIAL AND METHODS 3.1 NIRS METHODOLOGY AND SPECTRA COLLECTION 07 3.2 CRITERIA FOR SELECTING XYLARIA TO BE VISITED TO OBTAIN SPECTRAS 07 3 3 TERMINOLOGY 08 4. RESULTS 4.1 CONTACTED XYLARIA FOR COLLECTION SURVEY 10 4.1.1 BRAZILIAN XYLARIA 10 4.1.2 INTERNATIONAL XYLARIA 11 4.2 SELECTED XYLARIA 11 4.3 RESULTS OF THE SURVEY OF DALBERGIA SAMPLES IN THE BRAZILIAN XYLARIA 13 4.4 RESULTS OF THE SURVEY OF DALBERGIA SAMPLES IN THE INTERNATIONAL XYLARIA 14 5. CONCLUSION AND RECOMMENDATIONS 19 6. REFERENCES 20 APPENDICES 22 APPENDIX I DALBERGIA IN BRAZILIAN XYLARIA 22 CACAO RESEARCH CENTER – CEPECw 22 EMÍLIO GOELDI MUSEUM – M. -

International Journal of Scientific Research and Reviews
Kumar B. Sunil et al., IJSRR 2018, 7(3), 1968-1972 Review article Available online www.ijsrr.org ISSN: 2279–0543 International Journal of Scientific Research and Reviews The Rapeutic Properties of Red Sandal Wood- A Review * ** Kumar B. Sunil , Kumar T. Ganesh * Faculty & Head, Department of Botany, CSSR&SRRM Degree & PG College, Kamalapuram,YSR Kadapa Dist. A.P., Mobile: 8374790219, Email: [email protected] ** Faculty & Head, Department of Chemistry, CSSR&SRRM Degree & PG College, Kamalapuram,YSR Kadapa Dist. A.P., Mobile: 9000724247, Email: [email protected] ABSTRACT Pterocarpus santalinus, also known as „red sanders ‟ or „red sandalwood‟ is a highly valuable forest legume tree. It is locally known as „Rakta Chandan‟. This species occurs utterly in a well- defined forest area of Andhra Pradesh in Southern India. Now included in red list of endangered plants under IUCN guidelines. It contains many other compounds that have medicinal properties. Since the beginning of civilization in sub-continent, this plant is widely used in „Ayurved‟ in India. In recent years different studies showed the antimicrobial activity of the leaf extracts, stem bark extracts‟ from this plant. This review paper discusses the therapeutic properties of Red sandal wood. KEYWORDS: Red sandal wood, antimicrobial activity, anti-ageing agent. *Corresponding author B. Sunil Kumar Faculty & Head, Department of Botany, CSSR&SRRM Degree & PG College, Kamalapuram,YSR Kadapa Dist. A.P., Mobile: 8374790219, Email: [email protected] IJSRR, 7(3) July – Sep., 2018 Page 1968 Kumar B. Sunil et al., IJSRR 2018, 7(3), 1968-1972 INTRODUCTION Red Sandalwood is a species of Pterocarpus native to India. -

Analyse De La Croissance En Épaisseur De Dalbergia Baronii (Palissandre) Et De Dalbergia Monticola (Bois De Rose) Dans La Forê
Analyse de la croissance en épaisseur de Dalbergia baronii (palissandre) et de Dalbergia monticola (bois de rose) dans la forêt classée d’Ambohilero sous transfert de gestion Felana Niaina Rakoto Joseph, Bako Harisoa Ravaomanalina, Fenonirina Rakotoarison, Edmond Roger, Bakolimalala Rakouth To cite this version: Felana Niaina Rakoto Joseph, Bako Harisoa Ravaomanalina, Fenonirina Rakotoarison, Edmond Roger, Bakolimalala Rakouth. Analyse de la croissance en épaisseur de Dalbergia baronii (palissandre) et de Dalbergia monticola (bois de rose) dans la forêt classée d’Ambohilero sous transfert de gestion. Rôle et place des transferts de gestion des ressources naturelles renouvelables dans les politiques forestières actuelles à Madagascar, Dec 2013, France. pp.4. cirad-00933719 HAL Id: cirad-00933719 http://hal.cirad.fr/cirad-00933719 Submitted on 21 Jan 2014 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Analyse de la croissance en épaisseur de Dalbergia baronii (palissandre) et de Dalbergia monticola (bois de rose) dans la forêt classée d’Ambohilero sous transfert de gestion RAKOTO JOSEPH Felana Niaina1, RAVAOMANALINA Bako Harisoa1, RAKOTOARISON Fenonirina1, ROGER Edmond1 et RAKOUTH Bakolimalala1. 1 Département de biologie et écologie végétales, Faculté des sciences, Université d’Antananarivo, BP 906, Antananarivo 101, Madagascar. -
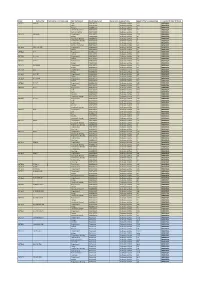
List of Instruments Affected by CITES II
Brand Itemnumber Itemnumber incl. -

Precious Woods Background Paper 1
Chatham House Workshop: Tackling the Trade in Illegal Precious Woods 23-24 April 2012 Background Paper 1: Precious Woods: Exploitation of the Finest Timber Prepared by TRAFFIC Authors: Section 1: Anna Jenkins, Neil Bridgland, Rachel Hembery & Ulrich Malessa Section 2: James Hewitt, Ulrich Malessa & Chen Hin Keong This review was commissioned from TRAFFIC by The Royal Institute of International Affairs (Chatham House), London UK. TRAFFIC supervised the elaboration of the review with support of Ethical Change Ltd, Llanidloes UK. The review was developed as one of three studies to explore the social and ecological impacts of trade, related exporting and importing country regulations as well as to develop recommendations to reduce the negative impacts of trade in precious woods species. Contact details of lead authors and supervisor: Section 1 & Appendices Anna Jenkins Ethical Change Ltd Tryfan, Llanidloes, SY18 6HU, Wales, UK [email protected] Section 2 James Hewitt [email protected] Section 1 & 2 (technical supervisor) Ulrich Malessa TRAFFIC WWF US 1250 24 th ST NW, Washington, DC 20037, USA [email protected] 2 Contents Contents ............................................................................................................................................................................................. 3 Acknowledgments ....................................................................................................................................................................... 4 Section 1 ............................................................................................................................................................................................ -

Wood Toxicity: Symptoms, Species, and Solutions by Andi Wolfe
Wood Toxicity: Symptoms, Species, and Solutions By Andi Wolfe Ohio State University, Department of Evolution, Ecology, and Organismal Biology Table 1. Woods known to have wood toxicity effects, arranged by trade name. Adapted from the Wood Database (http://www.wood-database.com). A good reference book about wood toxicity is “Woods Injurious to Human Health – A Manual” by Björn Hausen (1981) ISBN 3-11-008485-6. Table 1. Woods known to have wood toxicity effects, arranged by trade name. Adapted from references cited in article. Trade Name(s) Botanical name Family Distribution Reported Symptoms Affected Organs Fabaceae Central Africa, African Blackwood Dalbergia melanoxylon Irritant, Sensitizer Skin, Eyes, Lungs (Legume Family) Southern Africa Meliaceae Irritant, Sensitizer, African Mahogany Khaya anthotheca (Mahogany West Tropical Africa Nasopharyngeal Cancer Skin, Lungs Family) (rare) Meliaceae Irritant, Sensitizer, African Mahogany Khaya grandifoliola (Mahogany West Tropical Africa Nasopharyngeal Cancer Skin, Lungs Family) (rare) Meliaceae Irritant, Sensitizer, African Mahogany Khaya ivorensis (Mahogany West Tropical Africa Nasopharyngeal Cancer Skin, Lungs Family) (rare) Meliaceae Irritant, Sensitizer, African Mahogany Khaya senegalensis (Mahogany West Tropical Africa Nasopharyngeal Cancer Skin, Lungs Family) (rare) Fabaceae African Mesquite Prosopis africana Tropical Africa Irritant Skin (Legume Family) African Padauk, Fabaceae Central and Tropical Asthma, Irritant, Nausea, Pterocarpus soyauxii Skin, Eyes, Lungs Vermillion (Legume Family) -

First Molecular Phylogeny of the Pantropical Genus Dalbergia: Implications for Infrageneric Circumscription and Biogeography
SAJB-00970; No of Pages 7 South African Journal of Botany xxx (2013) xxx–xxx Contents lists available at SciVerse ScienceDirect South African Journal of Botany journal homepage: www.elsevier.com/locate/sajb First molecular phylogeny of the pantropical genus Dalbergia: implications for infrageneric circumscription and biogeography Mohammad Vatanparast a,⁎, Bente B. Klitgård b, Frits A.C.B. Adema c, R. Toby Pennington d, Tetsukazu Yahara e, Tadashi Kajita a a Department of Biology, Graduate School of Science, Chiba University, 1-33 Yayoi, Inage, Chiba, Japan b Herbarium, Library, Art and Archives, Royal Botanic Gardens, Kew, Richmond, United Kingdom c NHN Section, Netherlands Centre for Biodiversity Naturalis, Leiden University, Leiden, The Netherlands d Royal Botanic Garden Edinburgh, 20a Inverleith Row, Edinburgh, EH3 5LR, United Kingdom e Department of Biology, Kyushu University, Japan article info abstract Article history: The genus Dalbergia with c. 250 species has a pantropical distribution. In spite of the high economic and eco- Received 19 May 2013 logical value of the genus, it has not yet been the focus of a species level phylogenetic study. We utilized ITS Received in revised form 29 June 2013 nuclear sequence data and included 64 Dalbergia species representative of its entire geographic range to pro- Accepted 1 July 2013 vide a first phylogenetic framework of the genus to evaluate previous infrageneric classifications based on Available online xxxx morphological data. The phylogenetic analyses performed suggest that Dalbergia is monophyletic and that fi Edited by JS Boatwright it probably originated in the New World. Several clades corresponding to sections of these previous classi - cations are revealed. -

Table De Matiere
UNIVERSITE D'ANTANANARIVO DOMAINE DES SCIENCES ET TECHNOLOGIES ECOLE DOCTORALE EN SCIENCES DE LA VIE ET DE L’ENVIRONNEMENT THESE POUR L'OBTENTION DU DIPLOME DE DOCTORAT EN SCIENCES DE LA VIE ET DE L'ENVIRONNEMENT SPECIALITE : SCIENCES DU VEGETAL APPORTS DES ESPECES PIONNIERES ET DES PROPRIETES DU SOL A LA REGENERATION FORESTIERE DANS LE CORRIDOR FORESTIER FANDRIANA-VONDROZO Présentée par Andry RANDRIANARISON Soutenue publiquement, le 10 janvier 2017 devant le jury composé de : Président : Professeur Miadana Harisoa FARAMALALA Directeur de thèse : Professeur Vonjison RAKOTOARIMANANA Co-directeur de thèse : Professeur Alexandre BUTTLER Rapporteur interne: Professeur Bakolimalala RAKOUTH Rapporteur externe : Professeur Samuel RAZANAKA Examinateur : Professeur Josoa R. RANDRIAMALALA Invité : Docteur Dominique HERVÉ Laboratoire des systèmes écologiques (ECOS) UNIVERSITE D'ANTANANARIVO DOMAINE DES SCIENCES ET TECHNOLOGIES ECOLE DOCTORALE EN SCIENCES DE LA VIE ET DE L’ENVIRONNEMENT THESE POUR L'OBTENTION DU DIPLOME DE DOCTORAT EN SCIENCES DE LA VIE ET DE L'ENVIRONNEMENT SPECIALITE : SCIENCES DU VEGETAL APPORTS DES ESPECES PIONNIERES ET DES PROPRIETES DU SOL A LA REGENERATION FORESTIERE DANS LE CORRIDOR FORESTIER FANDRIANA-VONDROZO Présentée par Andry RANDRIANARISON Soutenue publiquement, le 10 janvier 2017 devant le jury composé de : Président : Professeur Miadana Harisoa FARAMALALA Directeur de thèse : Professeur Vonjison RAKOTOARIMANANA Co-directeur de thèse : Professeur Alexandre BUTTLER Rapporteur interne: Professeur Bakolimalala RAKOUTH Rapporteur externe : Professeur Samuel RAZANAKA Examinateur : Professeur Josoa R. RANDRIAMALALA Invité : Docteur Dominique HERVÉ Remerciements Il m’est agréable de présenter ma profonde gratitude et de remercier tous ceux qui m’ont formé, aidé, soutenu, accueilli, et de quelque façon m’ont permis de mener à bien cette recherche. -

CITES and Timber (PDF)
This guide covers the main timber species regulated CITES and Timber by the Convention on International Trade in Endangered Species (CITES). It provides information CITES and Timber on the key issues regarding the implementation of the Convention for this important group of plants. A guide to CITES-listed tree species Written for the non-expert, individual sections cover the species found in significant trade, with details on their distribution, uses, traded parts and derivatives, and scientific and common names. Madeleine Groves Madeleine Groves Additional sections cover timber identification and measurement, guidance on CITES documentation and key resources. and Catherine Rutherford shop.kew.org/kewbooksonline Madeleine Groves Catherine Rutherford CITES and Timber A guide to CITES-listed tree species Madeleine Groves Catherine Rutherford © The Board of Trustees of the Royal Botanic Gardens, Kew 2015 Illustrations and photographs © Royal Botanic Gardens, Kew, unless otherwise stated in the captions The authors have asserted their rights to be identified as the authors of this work in accordance with the Copyright, Designs and Patents Act 1988 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form, or by any means, electronic, mechanical, photocopying, recording or otherwise, without written permission of the publisher unless in accordance with the provisions of the Copyright Designs and Patents Act 1988. Great care has been taken to maintain the accuracy of the information contained in this work. However, neither the publisher, the editors nor authors can be held responsible for any consequences arising from use of the information contained herein. -

Trade in Dalbergia Nigra and the European Union
TRADE IN DALBERGIA NIGRA AND THE EUROPEAN UNION VICTORIA TAYLOR,KATALIN KECSE-NAGY AND THOMAS OSBORN A TRAFFIC REPORT Published by TRAFFIC. Report prepared by TRAFFIC for the European Commission under Contract 070307/2010/574210/SER/E2 © European Commission. All rights reserved. All material appearing in this publication is copyrighted and may be reproduced with permission. Any reproduction in full or in part of this publication must credit the European Commission as the copyright owner. The views of the authors expressed in this publication do not necessarily reflect those of the European Commission, TRAFFIC, WWF or IUCN. The designation of geographical entities in this publication, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of the European Commission, TRAFFIC or its supporting organizations concerning the legal status of any country, territory, or area, or its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership is held by WWF. TRAFFIC is a strategic alliance of WWF and IUCN. Suggested citation: Taylor, V., Kecse-Nagy, K. and Osborn, T. (2012). Trade in Dalbergia nigra and the European Union. Report prepared for the European Commission. ISBN 978-1-85850-355-4 Front cover photograph: Dalbergia nigra tree. Photograph credit: Mauricio Mercadante/Flickr.com, Creative Commons. Trade in Dalbergia nigra and the European Union June 2012 Victoria Taylor, Katalin Kecse-Nagy and Thomas Osborn A TRAFFIC Report prepared for the European Commission Version edited for public release Contract 070307/2010/574210/SER/E2 CONTENTS 1. Executive summary ................................................................................................................................... -

Cop16 Prop. 63
Original language: French CoP16 Prop. 63 CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________________ Sixteenth meeting of the Conference of the Parties Bangkok (Thailand), 3-14 March 2013 CONSIDERATION OF PROPOSALS FOR AMENDMENT OF APPENDICES I AND II A. Proposal Inclusion of the genus Dalbergia (populations of Madagascar) in CITES Appendix II: – In compliance with Article II, paragraph 2(a) of the Convention, and the Resolution Conf. 9.24 (Rev. CoP13), Annex 2 a, Paragraph A. – For similar reasons, in compliance with Article II, paragraph 2(a) of the Convention, and the Resolution Conf. 9.24 (Rev. CoP13), Annex 2 b, Paragraph A. We propose that the listing be limited to logs, sawn wood and veneer sheets and that the listing be annotated to that end, according to the recommendations of the Plants Committee (PC20, Dublin, 2012). B. Proponent Madagascar*. C. Supporting statement 1. Taxonomy 1.1 Class: Magnoliopsida 1.2 Order: Fabales 1.3 Family: Leguminosae (Fabaceae) Juss. 1789 1.4 Genus, species or subspecies, including author and year: Dalbergia Hemsley (Schatz, 2001) The list of accepted names of the Dalbergia species and their synonyms in the Catalogue des Plantes Vasculaires de Madagascar (Catalogue of the Vascular Plants of Madagascar) is provided in Annex 1. 1.5 Scientific synonyms: (see Annex 1) 1.6 Common names: French: In Madagascar, there are two categories of Dalbergia: bois de rose and palissandre. English: Rosewood, Palisander * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat or the United Nations Environment Programme concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries. -

Red Sanders in Rayalaseema Region of Andhra Pradesh: Importance to Commercial & Medicinal Value
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN:2278-3008, p-ISSN:2319-7676. Volume 11, Issue 1 Ver. IV (Jan.- Feb.2016), PP 57-60 www.iosrjournals.org Red Sanders in Rayalaseema Region of Andhra Pradesh: Importance to Commercial & Medicinal Value 1Dr. V. Ramabrahmam, 2Ms. Sujatha, (Ph.D) Assistant Professor Dept. of History & Archaeology Yogi Vemana University Kadapa – 516 003, A. P. Research Scholar Dept. of Anthropology S.V. University Tirupati - 517 502, A. P. Pterocarpus santalinus, with the common names Red Sanders, Red Sandalwood, and Saunders wood, is a species of Pterocarpus endemic to the southern Eastern Ghats mountain range of South India. This tree is valued for the rich red color of its wood. The wood is not aromatic. The tree is not to be confused with the aromatic Santalum Sandalwood trees that grow natively in South India. Red Sander (RS) is an endangered timber tree species, endemic to southern India. It grows in approximately 5160 km2 of fragmented forest landscape of southern Andhra Pradesh, and in a few sporadic patches in Tamil Nadu and Karnataka states. The wood is primarily used for making musical instruments and luxury furniture. It also yields Santa line dye which finds use in coloring foodstuff and pharmaceutical Preparations.1 CLASSIFICATION Genus Pterocarpus Species Pterocarpus Santalinus Kingdom Plantae (unranked) Angiosperms (unranked) Eudicots (unranked) Rosids Order Fabales Family Fabaceae Subfamily Faboideae Tribe Dalbergieae Other Names Red sanders, Red Sandalwood, Ruby wood Hindi Names Lal Chandan, Rakta Chandan It is a light-demanding moderate sized tree growing up to 8 m tall with a trunk 50–150 cm diameter.