Circovirus Infection in Cattle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Guide for Common Viral Diseases of Animals in Louisiana
Sampling and Testing Guide for Common Viral Diseases of Animals in Louisiana Please click on the species of interest: Cattle Deer and Small Ruminants The Louisiana Animal Swine Disease Diagnostic Horses Laboratory Dogs A service unit of the LSU School of Veterinary Medicine Adapted from Murphy, F.A., et al, Veterinary Virology, 3rd ed. Cats Academic Press, 1999. Compiled by Rob Poston Multi-species: Rabiesvirus DCN LADDL Guide for Common Viral Diseases v. B2 1 Cattle Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 2 Deer and Small Ruminants Please click on the principle system involvement Generalized viral disease Respiratory viral disease Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 3 Swine Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 4 Horses Please click on the principle system involvement Generalized viral diseases Neurological viral diseases Respiratory viral diseases Enteric viral diseases Abortifacient/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 5 Dogs Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. -

Encapsulation of Biomolecules in Bacteriophage MS2 Viral Capsids
Encapsulation of Biomolecules in Bacteriophage MS2 Viral Capsids By Jeff Edward Glasgow A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Chemistry in the Graduate Division of the University of California, Berkeley Committee in Charge: Professor Matthew Francis, Co-Chair Professor Danielle Tullman-Ercek, Co-Chair Professor Michelle Chang Professor John Dueber Spring 2014 Encapsulation of Biomolecules in Bacteriophage MS2 Viral Capsids Copyright ©2014 By: Jeff Edward Glasgow Abstract Encapsulation of Biomolecules in Bacteriophage MS2 Viral Capsids By Jeff Edward Glasgow Doctor of Philosophy in Chemistry University of California, Berkeley Professor Matthew Francis, Co-Chair Professor Danielle Tullman-Ercek, Co-Chair Nanometer-scale molecular assemblies have numerous applications in materials, catalysis, and medicine. Self-assembly has been used to create many structures, but approaches can match the extraordinary combination of stability, homogeneity, and chemical flexibility found in viral capsids. In particular, the bacteriophage MS2 capsid has provided a porous scaffold for several engineered nanomaterials for drug delivery, targeted cellular imaging, and photodynamic thera- py by chemical modification of the inner and outer surfaces of the shell. This work describes the development of new methods for reassembly of the capsid with concomitant encapsulation of large biomolecules. These methods were then used to encapsulate a variety of interesting cargoes, including RNA, DNA, protein-nucleic acid and protein-polymer conjugates, metal nanoparticles, and enzymes. To develop a stable, scalable method for encapsulation of biomolecules, the assembly of the capsid from its constituent subunits was analyzed in detail. It was found that combinations of negatively charged biomolecules and protein stabilizing agents could enhance reassembly, while electrostatic interactions of the biomolecules with the positively charged inner surface led to en- capsulation. -

Novel Circoviruses Detected in Feces of Sonoran Felids
viruses Article Novel Circoviruses Detected in Feces of Sonoran Felids Natalie Payne 1, Simona Kraberger 2, Rafaela S Fontenele 2, Kara Schmidlin 2, Melissa H Bergeman 3 , Ivonne Cassaigne 4, Melanie Culver 5,6,1, Arvind Varsani 2,7,* and Koenraad Van Doorslaer 1,3,8,* 1 Genetics Graduate Interdisciplinary Program, University of Arizona, Tucson, AZ 85719, USA; [email protected] 2 The Biodesign Center for Fundamental and Applied Microbiomics, Center for Evolution and Medicine, School of Life Sciences, Arizona State University, Tempe, AZ 85287-5001, USA; [email protected] (S.K.); [email protected] (R.S.F.); [email protected] (K.S.) 3 School of Animal and Comparative Biomedical Sciences, University of Arizona, Tucson, AZ 85721, USA; [email protected] 4 Primero Conservation, Box 16106, Portal, AZ 85632, USA; [email protected] 5 U.S. Geological Survey, Arizona Cooperative Fish and Wildlife Research Unit, University of Arizona, Tucson, AZ 85721, USA; [email protected] 6 School of Natural Resources and the Environment, University of Arizona, Tucson, AZ 85721, USA 7 Structural Biology Research Unit, Department of Integrative Biomedical Sciences, University of Cape Town, Observatory, Cape Town 7701, South Africa 8 The BIO5 Institute, Department of Immunobiology, Cancer Biology Graduate Interdisciplinary Program, UA Cancer Center, University of Arizona Tucson, Tucson, AZ 85724, USA * Correspondence: [email protected] (A.V.); [email protected] (K.V.D.) Received: 22 August 2020; Accepted: 10 September 2020; Published: 15 September 2020 Abstract: Sonoran felids are threatened by drought and habitat fragmentation. Vector range expansion and anthropogenic factors such as habitat encroachment and climate change are altering viral evolutionary dynamics and exposure. -

Virus–Host Interactions and Their Roles in Coral Reef Health and Disease
!"#$"%& Virus–host interactions and their roles in coral reef health and disease Rebecca Vega Thurber1, Jérôme P. Payet1,2, Andrew R. Thurber1,2 and Adrienne M. S. Correa3 !"#$%&'$()(*+%&,(%--.#(+''/%!01(1/$%0-1$23++%(#4&,,+5(5&$-%#6('+1#$0$/$-("0+708-%#0$9(&17( 3%+7/'$080$9(4+$#3+$#6(&17(&%-($4%-&$-1-7("9(&1$4%+3+:-10'(70#$/%"&1'-;(<40#(=-80-5(3%+807-#( &1(01$%+7/'$0+1($+('+%&,(%--.(80%+,+:9(&17(->34�?-#($4-(,01@#("-$5--1(80%/#-#6('+%&,(>+%$&,0$9( &17(%--.(-'+#9#$->(7-',01-;(A-(7-#'%0"-($4-(70#$01'$08-("-1$40'2&##+'0&$-7(&17(5&$-%2'+,/>12( &##+'0&$-7(80%+>-#($4&$(&%-(/10B/-($+('+%&,(%--.#6(540'4(4&8-(%-'-08-7(,-##(&$$-1$0+1($4&1( 80%/#-#(01(+3-12+'-&1(#9#$->#;(A-(493+$4-#0?-($4&$(80%/#-#(+.("&'$-%0&(&17(-/@&%9+$-#( 791&>0'&,,9(01$-%&'$(50$4($4-0%(4+#$#(01($4-(5&$-%('+,/>1(&17(50$4(#',-%&'$010&1(C#$+19D('+%&,#($+( 01.,/-1'-(>0'%+"0&,('+>>/10$9(791&>0'#6('+%&,(",-&'401:(&17(70#-&#-6(&17(%--.("0+:-+'4->0'&,( cycling. Last, we outline how marine viruses are an integral part of the reef system and suggest $4&$($4-(01.,/-1'-(+.(80%/#-#(+1(%--.(./1'$0+1(0#(&1(-##-1$0&,('+>3+1-1$(+.($4-#-(:,+"&,,9( 0>3+%$&1$(-180%+1>-1$#; To p - d ow n e f f e c t s Viruses infect all cellular life, including bacteria and evidence that macroorganisms play important parts in The ecological concept that eukaryotes, and contain ~200 megatonnes of carbon the dynamics of viroplankton; for example, sponges can organismal growth and globally1 — thus, they are integral parts of marine eco- filter and consume viruses6,7. -

25 May 7, 2014
Joint Pathology Center Veterinary Pathology Services Wednesday Slide Conference 2013-2014 Conference 25 May 7, 2014 ______________________________________________________________________________ CASE I: 3121206023 (JPC 4035610). Signalment: 5-week-old mixed breed piglet, (Sus domesticus). History: Two piglets from the faculty farm were found dead, and another piglet was weak and ataxic and, therefore, euthanized. Gross Pathology: The submitted piglet was in good body condition. It was icteric and had a diffusely pale liver. Additionally, petechial hemorrhages were found on the kidneys, and some fibrin was present covering the abdominal organs. Laboratory Results: The intestine was PCR positive for porcine circovirus (>9170000). Histopathologic Description: Mesenteric lymph node: Diffusely, there is severe lymphoid depletion with scattered karyorrhectic debris (necrosis). Also scattered throughout the section are large numbers of macrophages and eosinophils. The macrophages often contain botryoid basophilic glassy intracytoplasmic inclusion bodies. In fewer macrophages, intranuclear basophilic inclusions can be found. Liver: There is massive loss of hepatocytes, leaving disrupted liver lobules and dilated sinusoids engorged with erythrocytes. The remaining hepatocytes show severe swelling, with micro- and macrovesiculation of the cytoplasm and karyomegaly. Some swollen hepatocytes have basophilic intranuclear, irregular inclusions (degeneration). Throughout all parts of the liver there are scattered moderate to large numbers of macrophages (without inclusions). Within portal areas there is multifocally mild to moderate fibrosis and bile duct hyperplasia. Some bile duct epithelial cells show degeneration and necrosis, and there is infiltration of neutrophils within the lumen. The limiting plate is often obscured mainly by infiltrating macrophages and eosinophils, and fewer neutrophils, extending into the adjacent parenchyma. Scattered are small areas with extra medullary hematopoiesis. -

Canine Circovirus in Foxes from Northern Italy: Where Did It All Begin?
pathogens Article Canine Circovirus in Foxes from Northern Italy: Where Did It All Begin? Giovanni Franzo 1,* , Maria Luisa Menandro 1 , Claudia Maria Tucciarone 1 , Giacomo Barbierato 1, Lorenzo Crovato 1 , Alessandra Mondin 1, Martina Libanora 2, Federica Obber 2, Riccardo Orusa 3, Serena Robetto 3, Carlo Citterio 2 and Laura Grassi 1 1 Department of Animal Medicine, Production and Health (MAPS), University of Padua, 35020 Legnaro, Italy; [email protected] (M.L.M.); [email protected] (C.M.T.); [email protected] (G.B.); [email protected] (L.C.); [email protected] (A.M.); [email protected] (L.G.) 2 O.U. of Ecopathology, SCT2 Belluno, Istituto Zooprofilattico Sperimentale delle Venezie (IZSVe), 32100 Belluno, Italy; [email protected] (M.L.); [email protected] (F.O.); [email protected] (C.C.) 3 S.C. Valle d.’Aosta—National Reference Centre Wildlife Diseases, Istituto Zooprofilattico Sperimentale del Piemonte, Liguria e Valle d’Aosta (IZS PLV)—Ce.R.M.A.S., 11020 Quart, AO, Italy; [email protected] (R.O.); [email protected] (S.R.) * Correspondence: [email protected]; Tel.: +39-049-827-2968 Abstract: Canine circovirus (CanineCV) is a recently identified virus affecting both domestic and wild carnivores, including foxes, sometimes in presence of severe clinical signs. Its circulation in wild animals can thus represent a potential threat for endangered species conservation and an infection Citation: Franzo, G.; Menandro, source for dogs. Nevertheless, no data were available on its circulation in the Alps region of Northern M.L.; Tucciarone, C.M.; Barbierato, G.; Italy. -

Circovirus in Domestic and Wild Carnivores an Important
Virology 490 (2016) 69–74 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/yviro Circovirus in domestic and wild carnivores: An important opportunistic agent? Guendalina Zaccaria a, Daniela Malatesta a, Gabriella Scipioni a, Elisabetta Di Felice a, Marco Campolo b,c, Claudia Casaccia a, Giovanni Savini a, Daria Di Sabatino a, Alessio Lorusso a,n a Virology Unit, Istituto Zooprofilattico Sperimentale dell’Abruzzo e Molise (IZSAM), Teramo, Italy b Veterinary Practitioner Centro Veterinario Einaudi, Bari, Italy c Zoo delle Maitine, Pesco Sannita, Benevento, Italy article info abstract Article history: Circoviruses are relatively novel pathogens with increased importance in canids. In this study, we first Received 23 October 2015 screened the presence of dog circovirus (DogCV) by molecular methods from a total number of 389 Returned to author for revisions internal organ samples originating from 277 individuals of domestic dogs and wild animals including 17 December 2015 wolves, foxes and badgers. All the animals originated from Central-Southern Italy, specifically from Accepted 11 January 2016 Abruzzi and Molise regions, areas hosting several natural parks. DogCV was detected in 9/34 wolves Available online 3 February 2016 (P¼26.4%; IC 95%: 14.6–43.1%), 8/209 dogs (P¼3.8%; IC 95%: 1.9–7.3%), 0/24 foxes (P¼0%; IC 95%: 0– Keywords: 13.8%), 1/10 badgers (P¼10%; IC 95%: 1.79–40.4%). However, all DogCV positive animals were shown to be Dog circovirus infected at least by an additional key pathogen, including canine distemper virus (CDV) and canine Co-infection parvovirus type 2. -
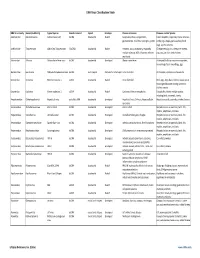
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics
viruses Article Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics Ashleigh F. Porter 1, Mang Shi 1, John-Sebastian Eden 1,2 , Yong-Zhen Zhang 3,4 and Edward C. Holmes 1,3,* 1 Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life & Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia; [email protected] (A.F.P.); [email protected] (M.S.); [email protected] (J.-S.E.) 2 Centre for Virus Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia 3 Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai 201500, China; [email protected] 4 Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China * Correspondence: [email protected]; Tel.: +61-2-9351-5591 Received: 17 October 2019; Accepted: 23 November 2019; Published: 25 November 2019 Abstract: DNA viruses comprise a wide array of genome structures and infect diverse host species. To date, most studies of DNA viruses have focused on those with the strongest disease associations. Accordingly, there has been a marked lack of sampling of DNA viruses from invertebrates. Bulk RNA sequencing has resulted in the discovery of a myriad of novel RNA viruses, and herein we used this methodology to identify actively transcribing DNA viruses in meta-transcriptomic libraries of diverse invertebrate species. Our analysis revealed high levels of phylogenetic diversity in DNA viruses, including 13 species from the Parvoviridae, Circoviridae, and Genomoviridae families of single-stranded DNA virus families, and six double-stranded DNA virus species from the Nudiviridae, Polyomaviridae, and Herpesviridae, for which few invertebrate viruses have been identified to date. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280