Altered Skeletal Muscle Mitochondrial Proteome As the Basis of Disruption of Mitochondrial Function in Diabetic Mice Piotr Zabie
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amplitaq and Amplitaq Gold DNA Polymerase
AmpliTaq and AmpliTaq Gold DNA Polymerase The Most Referenced Brand of DNA Polymerase in the World Date: 2005-05 Notes: Authors are listed alphabetically J. Biol. Chem. (223) Ezoe, S., I. Matsumura, et al. (2005). "GATA Transcription Factors Inhibit Cytokine-dependent Growth and Survival of a Hematopoietic Cell Line through the Inhibition of STAT3 Activity." J. Biol. Chem. 280(13): 13163-13170. http://www.jbc.org/cgi/content/abstract/280/13/13163 Although GATA-1 and GATA-2 were shown to be essential for the development of hematopoietic cells by gene targeting experiments, they were also reported to inhibit the growth of hematopoietic cells. Therefore, in this study, we examined the effects of GATA-1 and GATA-2 on cytokine signals. A tamoxifen-inducible form of GATA-1 (GATA-1/ERT) showed a minor inhibitory effect on interleukin-3 (IL-3)-dependent growth of an IL-3-dependent cell line Ba/F3. On the other hand, it drastically inhibited TPO-dependent growth and gp130-mediated growth/survival of Ba/F3. Similarly, an estradiol-inducible form of GATA-2 (GATA-2/ER) disrupted thrombopoietin (TPO)-dependent growth and gp130-mediated growth/survival of Ba/F3. As for this mechanism, we found that both GATA-1 and GATA-2 directly bound to STAT3 both in vitro and in vivo and inhibited its DNA-binding activity in gel shift assays and chromatin immunoprecipitation assays, whereas they hardly affected STAT5 activity. In addition, endogenous GATA-1 was found to interact with STAT3 in normal megakaryocytes, suggesting that GATA-1 may inhibit STAT3 activity in normal hematopoietic cells. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

The Purification and Identification of Interactors to Elucidate Novel Connections in the HEK 293 Cell Line
The Purification and Identification of Interactors to Elucidate Novel Connections in the HEK 293 Cell Line Brett Hawley Biochemistry, Microbiology and Immunology Faculty of Medicine University of Ottawa © Brett Hawley, Ottawa, Canada, 2012 ABSTRACT The field of proteomics studies the structure and function of proteins in a large scale and high throughput manner. My work in the field of proteomics focuses on identifying interactions between proteins and discovering novel interactions. The identification of these interactions provides new information on metabolic and disease pathways and the working proteome of a cell. Cells are lysed and purified using antibody based affinity purification followed by digestion and identification using an HPLC coupled to a mass spectrometer. In my studies, I looked at the interaction networks of several AD related genes (Apolipoprotein E, Clusterin variant 1 and 2, Low-density lipoprotein receptor, Phosphatidylinositol binding clathrin assembly protein, Alpha- synuclein and Platelet-activating factor receptor) and an endosomal recycling pathway involved in cholesterol metabolism (Eps15 homology domain 1,2 and 4, Proprotein convertase subtilisin/kexin type 9 and Low-density lipoprotein receptor). Several novel and existing interactors were identified and these interactions were validated using co-immunopurification, which could be the basis for future research. ii ACKNOWLEDGEMENTS I would like to take this opportunity to thank my supervisor, Dr. Daniel Figeys, for his support and guidance throughout my studies in his lab. It was a great experience to work in his lab and I am very thankful I was given the chance to learn and work under him. I would also like to thank the members of my lab for all their assistance in learning new techniques and equipment in the lab. -

CYP2A6) by P53
Transcriptional Regulation of Human Stress Responsive Cytochrome P450 2A6 (CYP2A6) by p53 Hao Hu M.Biotech. (Biotechnology) 2012 The University of Queensland B.B.A. 2009 University of Electronic Science and Technology of China B.Sc. (Pharmacy) 2009 University of Electronic Science and Technology of China A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2016 School of Medicine ABSTRACT Human cytochrome P450 (CYP) 2A6 is highly expressed in the liver and the encoding gene is regulated by various stress activated transcription factors, such as the nuclear factor (erythroid-derived 2)-like 2 (Nrf-2). Unlike the other xenobiotic metabolising CYP enzymes (XMEs), CYP2A6 only plays a minor role in xenobiotic metabolism. The CYP2A6 is highly induced by multiple forms of cellular stress conditions, where XMEs expression is normally inhibited. Recent findings suggest that the CYP2A6 plays an important role in regulating BR homeostasis. A computer based sequence analysis on the 3 kb proximate CYP2A6 promoter revealed several putative binding sites for p53, a protein that mediates regulation of antioxidant and apoptosis pathways. In this study, the role of p53 in CYP2A6 gene regulation is demonstrated. The site closest to transcription start site (TSS) is highly homologous with the p53 consensus sequence. The p53 responsiveness of this site was confirmed by transfections with various stepwise deleted of CYP2A6-5’-Luc constructs containing the putative p53RE. Deletion of the putative p53RE resulted in a total abolishment of p53 responsiveness of CYP2A6 promoter. Specific binding of p53 to the putative p53RE was detected by electrophoresis mobility shift assay. -

Inborn Errors of Metabolism Test Requisition
LABORATORY OF GENETICS AND GENOMICS Mailing Address: For local courier service and/or inquiries, please contact 513-636-4474 • Fax: 513-636-4373 3333 Burnet Avenue, Room R1042 www.cincinnatichildrens.org/moleculargenetics • Email: [email protected] Cincinnati, OH 45229 INBORN ERRORS OF METABOLISM TEST REQUISITION All Information Must Be Completed Before Sample Can Be Processed PATIENT INFORMATION ETHNIC/RACIAL BACKGROUND (Choose All) Patient Name: ___________________ , ___________________ , ________ European American (White) African-American (Black) Last First MI Native American or Alaskan Asian-American Address: ____________________________________________________ Pacific Islander Ashkenazi Jewish ancestry ____________________________________________________ Latino-Hispanic _____________________________________________ Home Phone: ________________________________________________ (specify country/region of origin) MR# __________________ Date of Birth ________ / ________ / _______ Other ____________________________________________________ (specify country/region of origin) Gender: Male Female BILLING INFORMATION (Choose ONE method of payment) o REFERRING INSTITUTION o COMMERCIAL INSURANCE* Insurance can only be billed if requested at the time of service. Institution: ____________________________________________________ Policy Holder Name: _____________________________________________ Address: _____________________________________________________ Gender: ________________ Date of Birth ________ / ________ / _______ -

) (51) International Patent Classification: Columbia V5G 1G3
) ( (51) International Patent Classification: Columbia V5G 1G3 (CA). PANDEY, Nihar R.; 10209 A 61K 31/4525 (2006.01) C07C 39/23 (2006.01) 128A St, Surrey, British Columbia V3T 3E7 (CA). A61K 31/05 (2006.01) C07D 405/06 (2006.01) (74) Agent: ZIESCHE, Sonia et al.; Gowling WLG (Canada) A61P25/22 (2006.01) LLP, 2300 - 550 Burrard Street, Vancouver, British Colum¬ (21) International Application Number: bia V6C 2B5 (CA). PCT/CA2020/050165 (81) Designated States (unless otherwise indicated, for every (22) International Filing Date: kind of national protection av ailable) . AE, AG, AL, AM, 07 February 2020 (07.02.2020) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (25) Filing Language: English DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, (26) Publication Language: English HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (30) Priority Data: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 16/270,389 07 February 2019 (07.02.2019) US OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (63) Related by continuation (CON) or continuation-in-part SC, SD, SE, SG, SK, SL, ST, SV, SY, TH, TJ, TM, TN, TR, (CIP) to earlier application: TT, TZ, UA, UG, US, UZ, VC, VN, WS, ZA, ZM, ZW. US 16/270,389 (CON) (84) Designated States (unless otherwise indicated, for every Filed on 07 Februaiy 2019 (07.02.2019) kind of regional protection available) . -

Genes Investigated
BabyNEXTTM EXTENDED Investigated genes and associated diseases Gene Disease OMIM OMIM Condition RUSP gene Disease ABCC8 Familial hyperinsulinism 600509 256450 Metabolic disorder - ABCC8-related Inborn error of amino acid metabolism ABCD1 Adrenoleukodystrophy 300371 300100 Miscellaneous RUSP multisystem (C) * diseases ABCD4 Methylmalonic aciduria and 603214 614857 Metabolic disorder - homocystinuria, cblJ type Inborn error of amino acid metabolism ACAD8 Isobutyryl-CoA 604773 611283 Metabolic Disorder - RUSP dehydrogenase deficiency Inborn error of (S) ** organic acid metabolism ACAD9 acyl-CoA dehydrogenase-9 611103 611126 Metabolic Disorder - (ACAD9) deficiency Inborn error of fatty acid metabolism ACADM Acyl-CoA dehydrogenase, 607008 201450 Metabolic Disorder - RUSP medium chain, deficiency of Inborn error of fatty (C) acid metabolism ACADS Acyl-CoA dehydrogenase, 606885 201470 Metabolic Disorder - RUSP short-chain, deficiency of Inborn error of fatty (S) acid metabolism ACADSB 2-methylbutyrylglycinuria 600301 610006 Metabolic Disorder - RUSP Inborn error of (S) organic acid metabolism ACADVL very long-chain acyl-CoA 609575 201475 Metabolic Disorder - RUSP dehydrogenase deficiency Inborn error of fatty (C) acid metabolism ACAT1 Alpha-methylacetoacetic 607809 203750 Metabolic Disorder - RUSP aciduria Inborn error of (C) organic acid metabolism ACSF3 Combined malonic and 614245 614265 Metabolic Disorder - methylmalonic aciduria Inborn error of organic acid metabolism 1 ADA Severe combined 608958 102700 Primary RUSP immunodeficiency due -

Dependent Traits in Mice Models of Hyperthyroidism and Hypothyroidism
Phenotypical characterization of sex- dependent traits in mice models of hyperthyroidism and hypothyroidism Inaugural-Dissertation zur Erlangung des Doktorgrades Dr. rer. nat. der Fakultät für Biologie an der Universität Duisburg-Essen vorgelegt von Helena Rakov aus St. Petropawlowsk April 2017 Die der vorliegenden Arbeit zugrunde liegenden Experimente wurden am Universitätsklinikum Essen in der Klinik für Endokrinologie und Stoffwechselerkrankungen durchgeführt. 1. Gutachter: Prof. Dr. Dr. Dagmar Führer-Sakel 2. Gutachter: Prof. Dr. Elke Cario Vorsitzender des Prüfungsausschusses: Prof. Dr. Ruth Grümmer Tag der mündlichen Prüfung: 17.07.2017 Publications Publications Engels Kathrin*, Rakov Helena *, Zwanziger Denise, Moeller Lars C., Homuth Georg, Köhrle Josef, Brix Klaudia, Fuhrer Dagmar. Differences in mouse hepatic thyroid hormone transporter expression with age and hyperthyroidism. Eur Thyroid J 2015;4(suppl 1):81–86. DOI: 10.1159/000381020. *contributed equally Zwanziger Denise*, Rakov Helena*, Engels Kathrin, Moeller Lars C., Fuhrer Dagmar. Sex-dependent claudin-1 expression in liver of eu- and hypothyroid mice. Eur Thyroid J. 2015 Sep; 4(Suppl 1): 67–73. DOI: 10.1159/000431316. *contributed equally Engels Kathrin*, Rakov Helena*, Zwanziger Denise, Hoenes Georg Sebastian, Rehders Maren, Brix Klaudia, Koehrle Josef, Moeller Lars Christian, Fuhrer Dagmar. Efficacy of protocols for induction of chronic hyperthyroidism in male and female mice. Endocrine. 2016 Oct;54(1):47-54. DOI: 10.1007/s12020-016-1020-8. Rakov Helena*, Engels Kathrin*, Hönes Georg Sebastian, Strucksberg Karl-Heinz, Moeller Lars Christian, Köhrle Josef, Zwanziger Denise, Führer Dagmar. Sex-specific phenotypes of hyperthyroidism and hypothyroidism in mice. Biol Sex Differ. 2016 Aug 24;7(1):36. DOI: 10.1186/s13293-016-0089-3. -

Deciphering the Gene Expression Profile of Peroxisome Proliferator
Chen et al. J Transl Med (2016) 14:157 DOI 10.1186/s12967-016-0871-3 Journal of Translational Medicine RESEARCH Open Access Deciphering the gene expression profile of peroxisome proliferator‑activated receptor signaling pathway in the left atria of patients with mitral regurgitation Mien‑Cheng Chen1*, Jen‑Ping Chang2, Yu‑Sheng Lin3, Kuo‑Li Pan3, Wan‑Chun Ho1, Wen‑Hao Liu1, Tzu‑Hao Chang4, Yao‑Kuang Huang5, Chih‑Yuan Fang1 and Chien‑Jen Chen1 Abstract Background: Differentially expressed genes in the left atria of mitral regurgitation (MR) pigs have been linked to peroxisome proliferator-activated receptor (PPAR) signaling pathway in the KEGG pathway. However, specific genes of the PPAR signaling pathway in the left atria of MR patients have never been explored. Methods: This study enrolled 15 MR patients with heart failure, 7 patients with aortic valve disease and heart failure, and 6 normal controls. We used PCR assay (84 genes) for PPAR pathway and quantitative RT-PCR to study specific genes of the PPAR pathway in the left atria. Results: Gene expression profiling analysis through PCR assay identified 23 genes to be differentially expressed in the left atria of MR patients compared to normal controls. The expressions of APOA1, ACADM, FABP3, ETFDH, ECH1, CPT1B, CPT2, SLC27A6, ACAA2, SMARCD3, SORBS1, EHHADH, SLC27A1, PPARGC1B, PPARA and CPT1A were significantly up-regulated, whereas the expression of PLTP was significantly down-regulated in the MR patients compared to normal controls. The expressions of HMGCS2, ACADM, FABP3, MLYCD, ECH1, ACAA2, EHHADH, CPT1A and PLTP were significantly up-regulated in the MR patients compared to patients with aortic valve disease. -
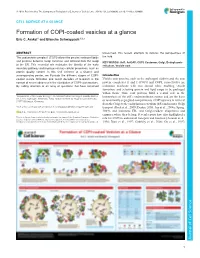
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Genetics of Migraine: Insights Into the Molecular Basis of Migraine Disorders
This may be the author’s version of a work that was submitted/accepted for publication in the following source: Sutherland, Heidi& Griffiths, Lyn (2017) Genetics of migraine: Insights into the molecular basis of migraine disor- ders. Headache, 57(4), pp. 537-569. This file was downloaded from: https://eprints.qut.edu.au/105633/ c Consult author(s) regarding copyright matters This work is covered by copyright. Unless the document is being made available under a Creative Commons Licence, you must assume that re-use is limited to personal use and that permission from the copyright owner must be obtained for all other uses. If the docu- ment is available under a Creative Commons License (or other specified license) then refer to the Licence for details of permitted re-use. It is a condition of access that users recog- nise and abide by the legal requirements associated with these rights. If you believe that this work infringes copyright please provide details by email to [email protected] Notice: Please note that this document may not be the Version of Record (i.e. published version) of the work. Author manuscript versions (as Sub- mitted for peer review or as Accepted for publication after peer review) can be identified by an absence of publisher branding and/or typeset appear- ance. If there is any doubt, please refer to the published source. https://doi.org/10.1111/head.13053 Genetics of Migraine: insights into the molecular basis of migraine disorders Heidi G. Sutherland, PhD and Lyn R. Griffiths, PhD Genomics Research Centre, Institute of Health and Biomedical Innovation, QUT, Musk Ave, Kelvin Grove, QLD 4059, Australia The authors declare no conflicts of interest.