Download (PDF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]](https://docslib.b-cdn.net/cover/8283/computational-genome-wide-identification-of-heat-shock-protein-genes-in-the-bovine-genome-version-1-peer-review-2-approved-1-approved-with-reservations-88283.webp)
Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]
F1000Research 2018, 7:1504 Last updated: 08 AUG 2021 RESEARCH ARTICLE Computational genome-wide identification of heat shock protein genes in the bovine genome [version 1; peer review: 2 approved, 1 approved with reservations] Oyeyemi O. Ajayi1,2, Sunday O. Peters3, Marcos De Donato2,4, Sunday O. Sowande5, Fidalis D.N. Mujibi6, Olanrewaju B. Morenikeji2,7, Bolaji N. Thomas 8, Matthew A. Adeleke 9, Ikhide G. Imumorin2,10,11 1Department of Animal Breeding and Genetics, Federal University of Agriculture, Abeokuta, Nigeria 2International Programs, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, 14853, USA 3Department of Animal Science, Berry College, Mount Berry, GA, 30149, USA 4Departamento Regional de Bioingenierias, Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, Queretaro, Mexico 5Department of Animal Production and Health, Federal University of Agriculture, Abeokuta, Nigeria 6Usomi Limited, Nairobi, Kenya 7Department of Animal Production and Health, Federal University of Technology, Akure, Nigeria 8Department of Biomedical Sciences, Rochester Institute of Technology, Rochester, NY, 14623, USA 9School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa 10School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, 30032, USA 11African Institute of Bioscience Research and Training, Ibadan, Nigeria v1 First published: 20 Sep 2018, 7:1504 Open Peer Review https://doi.org/10.12688/f1000research.16058.1 Latest published: 20 Sep 2018, 7:1504 https://doi.org/10.12688/f1000research.16058.1 Reviewer Status Invited Reviewers Abstract Background: Heat shock proteins (HSPs) are molecular chaperones 1 2 3 known to bind and sequester client proteins under stress. Methods: To identify and better understand some of these proteins, version 1 we carried out a computational genome-wide survey of the bovine 20 Sep 2018 report report report genome. -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

PLXNB1 (Plexin
Atlas of Genetics and Cytogenetics in Oncology and Haematology OPEN ACCESS JOURNAL AT INIST-CNRS Gene Section Mini Review PLXNB1 (plexin B1) José Javier Gómez-Román, Montserrat Nicolas Martínez, Servando Lazuén Fernández, José Fernando Val-Bernal Department of Anatomical Pathology, Marques de Valdecilla University Hospital, Medical Faculty, University of Cantabria, Santander, Spain (JJGR, MN, SL, JFVB) Published in Atlas Database: March 2009 Online updated version: http://AtlasGeneticsOncology.org/Genes/PLXNB1ID43413ch3p21.html DOI: 10.4267/2042/44702 This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2010 Atlas of Genetics and Cytogenetics in Oncology and Haematology Identity Pseudogene No. Other names: KIAA0407; MGC149167; OTTHUMP00000164806; PLEXIN-B1; PLXN5; SEP Protein HGNC (Hugo): PLXNB1 Location: 3p21.31 Description Local order: The Plexin B1 gene is located between 2135 Amino acids (AA). Plexins are receptors for axon FBXW12 and CCDC51 genes. molecular guidance molecules semaphorins. Plexin signalling is important in pathfinding and patterning of both neurons and developing blood vessels. Plexin-B1 is a surface cell receptor. When it binds to its ligand SEMA4D it activates several pathways by binding of cytoplasmic ligands, like RHOA activation and subsequent changes of the actin cytoskeleton, axon guidance, invasive growth and cell migration. It monomers and heterodimers with PLXNB2 after proteolytic processing. Binds RAC1 that has been activated by GTP binding. It binds PLXNA1 and by similarity ARHGEF11, Note ARHGEF12, ERBB2, MET, MST1R, RND1, NRP1 Size: 26,200 bases. and NRP2. Orientation: minus strand. This family features the C-terminal regions of various plexins. The cytoplasmic region, which has been called DNA/RNA a SEX domain in some members of this family is involved in downstream signalling pathways, by Description interaction with proteins such as Rac1, RhoD, Rnd1 and other plexins. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

De Novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation
bioRxiv preprint doi: https://doi.org/10.1101/757039; this version posted September 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. De novo EIF2AK1 and EIF2AK2 variants are associated with developmental delay, leukoencephalopathy, and neurologic decompensation Dongxue Mao1,2, Chloe M. Reuter3,4, Maura R.Z. Ruzhnikov5,6, Anita E. Beck7, Emily G. Farrow8,9,10, Lisa T. Emrick1,11,12,13, Jill A. Rosenfeld12, Katherine M. Mackenzie5, Laurie Robak2,12,13, Matthew T. Wheeler3,14, Lindsay C. Burrage12,13, Mahim Jain15, Pengfei Liu12, Daniel Calame11,13, Sebastien Küry17,18, Martin Sillesen19, Klaus Schmitz-Abe20, Davide Tonduti21, Luigina Spaccini22, Maria Iascone23, Casie A. Genetti20, Madeline Graf16, Alyssa Tran12, Mercedes Alejandro12, Undiagnosed Diseases Network, Brendan H. Lee12,13, Isabelle Thiffault8,9,24, Pankaj B. Agrawal#,20, Jonathan A. Bernstein#,3,25, Hugo J. Bellen#,2,12,26,27,28, Hsiao- Tuan Chao#,1,2,11,12,13,28,27,29 #Correspondence should be addressed: [email protected] (P.A.), [email protected] (J.A.B.), [email protected] (H.J.B.), [email protected] (H.T.C.) 1Department of Pediatrics, Baylor College of Medicine (BCM), Houston, TX 2Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 3Stanford Center for Undiagnosed Diseases, Stanford University, Stanford, CA 4Stanford Center for Inherited Cardiovascular Disease, Division of Cardiovascular Medicine, -

G-Protein-Coupled Receptor Signaling and Polarized Actin Dynamics Drive
RESEARCH ARTICLE elifesciences.org G-protein-coupled receptor signaling and polarized actin dynamics drive cell-in-cell invasion Vladimir Purvanov, Manuel Holst, Jameel Khan, Christian Baarlink, Robert Grosse* Institute of Pharmacology, University of Marburg, Marburg, Germany Abstract Homotypic or entotic cell-in-cell invasion is an integrin-independent process observed in carcinoma cells exposed during conditions of low adhesion such as in exudates of malignant disease. Although active cell-in-cell invasion depends on RhoA and actin, the precise mechanism as well as the underlying actin structures and assembly factors driving the process are unknown. Furthermore, whether specific cell surface receptors trigger entotic invasion in a signal-dependent fashion has not been investigated. In this study, we identify the G-protein-coupled LPA receptor 2 (LPAR2) as a signal transducer specifically required for the actively invading cell during entosis. We find that 12/13G and PDZ-RhoGEF are required for entotic invasion, which is driven by blebbing and a uropod-like actin structure at the rear of the invading cell. Finally, we provide evidence for an involvement of the RhoA-regulated formin Dia1 for entosis downstream of LPAR2. Thus, we delineate a signaling process that regulates actin dynamics during cell-in-cell invasion. DOI: 10.7554/eLife.02786.001 Introduction Entosis has been described as a specialized form of homotypic cell-in-cell invasion in which one cell actively crawls into another (Overholtzer et al., 2007). Frequently, this occurs between tumor cells such as breast, cervical, or colon carcinoma cells and can be triggered by matrix detachment (Overholtzer et al., 2007), suggesting that loss of integrin-mediated adhesion may promote cell-in-cell invasion. -

Mitochondrial Protein Quality Control Mechanisms
G C A T T A C G G C A T genes Review Mitochondrial Protein Quality Control Mechanisms Pooja Jadiya * and Dhanendra Tomar * Center for Translational Medicine, Lewis Katz School of Medicine, Temple University, Philadelphia, PA 19140, USA * Correspondence: [email protected] (P.J.); [email protected] (D.T.); Tel.: +1-215-707-9144 (D.T.) Received: 29 April 2020; Accepted: 15 May 2020; Published: 18 May 2020 Abstract: Mitochondria serve as a hub for many cellular processes, including bioenergetics, metabolism, cellular signaling, redox balance, calcium homeostasis, and cell death. The mitochondrial proteome includes over a thousand proteins, encoded by both the mitochondrial and nuclear genomes. The majority (~99%) of proteins are nuclear encoded that are synthesized in the cytosol and subsequently imported into the mitochondria. Within the mitochondria, polypeptides fold and assemble into their native functional form. Mitochondria health and integrity depend on correct protein import, folding, and regulated turnover termed as mitochondrial protein quality control (MPQC). Failure to maintain these processes can cause mitochondrial dysfunction that leads to various pathophysiological outcomes and the commencement of diseases. Here, we summarize the current knowledge about the role of different MPQC regulatory systems such as mitochondrial chaperones, proteases, the ubiquitin-proteasome system, mitochondrial unfolded protein response, mitophagy, and mitochondria-derived vesicles in the maintenance of mitochondrial proteome and health. The proper understanding of mitochondrial protein quality control mechanisms will provide relevant insights to treat multiple human diseases. Keywords: mitochondria; proteome; ubiquitin; proteasome; chaperones; protease; mitophagy; mitochondrial protein quality control; mitochondria-associated degradation; mitochondrial unfolded protein response 1. Introduction Mitochondria are double membrane, dynamic, and semiautonomous organelles which have several critical cellular functions. -
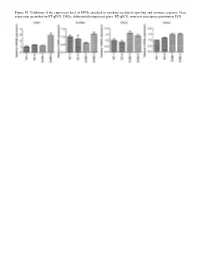
Figure S1. Validation of the Expression Level of Degs Enriched in Cytokine‑Mediated Signaling and Immune Response
Figure S1. Validation of the expression level of DEGs enriched in cytokine‑mediated signaling and immune response. Gene expression quantified by RT‑qPCR. DEGs, differentially expressed genes; RT‑qPCR, reverse‑transcription quantitative PCR. Figure S2. Validation of STAU1‑regulated AS events. IGV‑Sashimi plot revealed (A‑C) three A5SS AS events in three different genes. Reads distribution of each AS event was plotted in the left panel with the transcripts of each gene shown below. The sche‑ matic diagrams depict the structures of two AS events, AS1 (purple line) and AS2 (green line). The exon sequences are denoted by black boxes, the intron sequences by a horizontal line (right panel). RNA‑seq quantification and RT‑qPCR validation of ASEs are presented in the panels on the right. STAU1, double‑stranded RNA‑binding protein Staufen homolog 1; AS, alternative splicing; A5SS, alternative 5'splice site; RNA‑seq, RNA sequencing; RT‑qPCR, reverse‑transcription quantitative PCR. Error bars represent mean ± SEM. *P<0.05. Table SI. Primers used in gene validation experiments. IFIT2‑F CAGCCTACGGCAACTAAA IFIT2‑R GAGCCTTCTCAAAGCACA IFIT3‑F ACACCAAACAATGGCTAC IFIT3‑R TGGACAAACCCTCTAAAC OASL‑F AATGGTGACCGTGATGGG OASL‑R ACCTGAGGATGGAGCAGAG IFI27‑F TTCACTGCGGCGGGAATC IFI27‑R TGGCTGCTATGGAGGACGAG S1PR4‑F TGCTGAAGACGGTGCTGATG S1PR4‑R TGCGGAAGGAGTAGATGATGG CCL5‑F ACGACTGCTGGGTTGGAG CCL5‑R ACCCTGCTGCTTTGCCTA CCL2‑F CTAACCCAGAAACATCCAAT CCL2‑R GCTATGAGCAGCAGGCAC CD44‑F TGGAGGACAGAAAGCCAAGT CD44‑R TTCGCAATGAAACAATCAGTAG PLEKHG2‑M/As‑F CCAAAAGTAAGCCTGTCC PLEKHG2‑M‑R -

4 Understanding the Role of GNA13 Deregulation in Lymphomagenesis
Integrative Genomics Reveals a Role for GNA13 in Lymphomagenesis by Adrienne Greenough University Program in Genetics and Genomics Duke University Approved: ___________________________ Sandeep Dave, Supervisor ___________________________ Fred Dietrich ___________________________ Jack Keene ___________________________ Yuan Zhuang Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2014 i v ABSTRACT Integrative Genomics Reveals a Role for GNA13 in Lymphomagenesis by Adrienne Greenough University Program in Genetics and Genomics Duke University Approved: ___________________________ Sandeep Dave, Supervisor ___________________________ Fred Dietrich ___________________________ Jack Keene ___________________________ Yuan Zhuang An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2014 Copyright by Adrienne Greenough 2014 Abstract Lymphomas comprise a diverse group of malignancies derived from immune cells. High throughput sequencing has recently emerged as a powerful and versatile method for analysis of the cancer genome and transcriptome. As these data continue to emerge, the crucial work lies in sorting through the wealth of information to hone in on the critical aspects that will give us a better understanding of biology and new insight for how to treat disease. Finding the important signals within these large data sets is one of the major challenges of next generation sequencing. In this dissertation, I have developed several complementary strategies to describe the genetic underpinnings of lymphomas. I begin with developing a better method for RNA sequencing that enables strand-specific total RNA sequencing and alternative splicing profiling in the same analysis. -

Mapping the Spatial Proteome of Metastatic Cells in Colorectal Cancer
Mapping the spatial proteome of metastatic cells in colorectal cancer Marta Mendes‡¶, Alberto Peláez-García‡¥, María López-Lucendo‡, Ruben A. Bartolomé‡, Eva Calviño‡, Rodrigo Barderas‡§**, J. Ignacio Casal‡** ‡Department of Cellular and Molecular Medicine, Centro de Investigaciones Biológicas (CIB-CSIC), Madrid, Spain §Instituto de Salud Carlos III. Majadahonda. Spain ¶Present address: Technische Universität Berlin, Germany ¥Present address: Department of Pathology, Hospital Universitario La Paz, IdiPAZ, Madrid, Spain Running title: Mapping the metastatic proteome Keywords: Spatial proteome, cell fractionation, SILAC, metastasis, colorectal cancer ** To whom correspondence should be addressed: Department of Cellular and Molecular Medicine, Centro de Investigaciones Biológicas (CIB-CSIC), Ramiro de Maeztu, 9, 28040 Madrid, Spain. Tel.: +34 918373112; E-mail: [email protected]. Instituto de Salud Carlos III, Majadahonda, Spain Carretera de Majadahonda - Pozuelo, Km. 2.200, 28220 Majadahonda, Madrid, Spain. Tel.: +34 913944155; E-mail: [email protected] 1 ABSTRACT Colorectal cancer (CRC) is the second deadliest cancer worldwide. Here, we aimed to study metastasis mechanisms using spatial proteomics in the KM12 cell model. Cells were SILAC-labeled and fractionated into five subcellular fractions corresponding to: cytoplasm, plasma, mitochondria and ER/golgi membranes, nuclear, chromatin-bound and cytoskeletal proteins and analyzed with high resolution mass spectrometry. We provide localization data of 4863 quantified proteins in the different subcellular fractions. A total of 1318 proteins with at least 1.5-fold change were deregulated in highly metastatic KM12SM cells respect to KM12C cells. The protein network organization, protein complexes and functional pathways associated to CRC metastasis was revealed with spatial resolution. Although 92% of the differentially expressed proteins showed the same deregulation in all subcellular compartments, a subset of 117 proteins (8%) showed opposite changes in different subcellular localizations.