Rnase Mitochondrial RNA Processing Correctly Cleaves a Novel R Loop at the Mitochondrial DNA Leading-Strand Origin of Replication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcription from the Second Heavy-Strand Promoter of Human Mtdna Is Repressed by Transcription Factor a in Vitro
Transcription from the second heavy-strand promoter of human mtDNA is repressed by transcription factor A in vitro Maria F. Lodeiroa, Akira Uchidaa, Megan Bestwickb, Ibrahim M. Moustafaa, Jamie J. Arnolda, Gerald S. Shadelb,c, and Craig E. Camerona,1 aDepartment of Biochemistry and Molecular Biology, Pennsylvania State University, University Park, PA 16802; bDepartment of Pathology, Yale University School of Medicine, New Haven, CT 06520; and cDepartment of Genetics, Yale University School of Medicine, New Haven, CT 06520 Edited* by Douglas C. Wallace, Center for Mitochondrial and Epigenomic Medicine (CMEM), Children’s Hospital of Philadelphia, Philadelphia, PA, and approved March 8, 2012 (received for review November 15, 2011) Cell-based studies support the existence of two promoters on the factor B2 (TFB2M). The long-standing paradigm was that TFAM heavy strand of mtDNA: heavy-strand promoter 1 (HSP1) and HSP2. binds to a site in the promoter upstream of the transcription start However, transcription from HSP2 has been reported only once in site and recruits a complex POLRMT/TFB2M by an interaction of a cell-free system, and never when recombinant proteins have been the carboxyl-terminal tail of TFAM with TFB2M (3). However, we used. Here, we document transcription from HSP2 using an in vitro recently showed that basal mitochondrial transcription is not ab- system of defined composition. An oligonucleotide template repre- solutely dependent on TFAM (4). This observation suggested that senting positions 596–685 of mtDNA was sufficient to observe tran- the two-component transcription system found in lower eukaryotes scription by the human mtRNA polymerase (POLRMT) that was had acquired an additional layer of regulation in mammals that is absolutely dependent on mitochondrial transcription factor B2 mediated by TFAM (4). -

The Role of Control Region Mitochondrial DNA Mutations in Cardiovascular Disease: Stroke and Myocardial Infarction
bioRxiv preprint doi: https://doi.org/10.1101/382374; this version posted August 1, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. The role of control region mitochondrial DNA mutations in cardiovascular disease: stroke and myocardial infarction Control region mitochondrial DNA mutations in cardiovascular disease Miriam Umbria1, Amanda Ramos1,2,3,4, Maria Pilar Aluja1¶*,#a and Cristina Santos1¶*,#a 1 Unitat d’Antropologia Biològica, Departament de Biologia Animal, Biologia Vegetal i Ecologia. Facultat de Biociències. Universitat Autònoma de Barcelona. Cerdanyola del Vallès, Barcelona, Spain; GREAB – Research Group in Biological Anthropology. 2 Faculdade de Ciências e Tecnologia, Universidade dos Açores (UAc), Ponta Delgada, Portugal 3 Instituto de Investigação e Inovação em Saúde (I3S), Universidade do Porto, Portugal 4 Instituto de Biologia Molecular e Celular (IBMC), Universidade do Porto, Porto, Portugal #a Current Address: Unitat d’Antropologia Biològica, Departament de Biologia Animal, Biologia Vegetal i Ecologia. Facultat de Biociències. Universitat Autònoma de Barcelona, Cerdanyola del Vallès, Spain ¶ These authors contributed equally to this work *Correspondence to: Cristina Santos and Maria Pilar Aluja Tel.: 34 93 5811503 Fax: 34 93 5811321 E-mail: [email protected] E-mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/382374; this version posted August 1, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Ribonuclease and Deoxyribonuclease Activities in Experimental and Human Tumors by the Histochemical Substrate Film Method*
Ribonuclease and Deoxyribonuclease Activities in Experimental and Human Tumors by the Histochemical Substrate Film Method* R. DAOUSTJANDHARUKOAMANOÕ (Laboratoires de Recherche, Institut du Cancer de Montréal,Hôpital Notre-Dame et Universitéde Montréal,Montréal,Canada) SUMMARY The ribonuclease and deoxyribonuclease activities of 65 experimental and human tu mors (32 different types) have been examined by histochemical substrate film methods. A same general pattern was obtained for the distribution of both nucleases in the various types of experimental and human tumors. The connective tissue stroma and the necrotic regions of the tumor masses showed various levels of nuclease activity, whereas the neoplastic cells showed no demonstrable activity. It appears that deficien cies in ribonuclease and deoxyribonuclease activities represent general properties of cancer cells. The possible significance of the losses of nuclease activities in carcinogenesis is dis cussed. Studies on nucleases by histochemical methods MATERIALS AND METHODS have shown that losses of ribonuclease (RNase) The experimental tumors used in the present and deoxyribonuclease (DNase) activities take study were mostly rat, mouse, and hamster trans- place in rat liver during azo-dye carcinogenesis (1, plantable tumors (see Table 1). The tumor-bearing 6). The loss of RNase activity is progressive and animals were obtained from commercial or private occurs before parenchymal cells become cancerous, sources, and the tumors were used as supplied or whereas the loss of DNase activity is abrupt and closely associated with the neoplastic transforma TABLE1 tion of parenchymal cells. EXPERIMENTALTUMORS If a loss of RNase or DNase activity plays an important role in tumor formation, the lack of SpeciesRat"""MouseHamsterTumorPrimary demonstrable nuclease activity observed in rat hepatomaNovikoff primary hepatomas should also be observed in a hepatomaWalker variety of tumors. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

Enhanced Phosphorylation of High-Mobility-Group Proteins In
Proc. Nati. Acad. Sci. USA Vol. 78, No. 4, pp. 2189-2193, April 1981 Biochemistry Enhanced phosphorylation of high-mobility-group proteins in nuclease-sensitive mononucleosomes from butyrate-treated HeLa cells (hyperphosphorylation/transcriptionally active chromatin/sodium butyrate) BEATRIZ LEVY-WILSON Department of Molecular Biology and Biochemistry, University of California, Irvine, California 92717 Communicated by James F. Bonner, December 29, 1980 ABSTRACT The protein composition of nucleosome fractions MATERIALS AND METHODS differing in their sensitivity to micrococcal nuclease and derived from butyrate-treated or untreated HeLa cells has been com- Cell Cultures. HeLa cells, line S3, were grown in monolay- pared. Most of the high-mobility-group-14 (HMG-14) and HMG- ers, either in T-150 culture flasks or in roller bottles, at 370C 17 content of HeLa cell chromatin is associated with those nu- in minimal essential medium (Flow Laboratories, Rockville, cleosomes that are preferentially sensitive to micrococcal nu- MD) supplemented with 7% bovine calfserum (KC Biologicals) clease. Furthermore, electrophoresis of these two HMG proteins and penicillin (100 units/ml) and streptomycin (100 Ag/ml) from the transcriptionally active chromatin fraction MN, of bu- (Sigma). In experiments involving sodium butyrate, this com- tyrate-treated cells resolves them into a series ofbands. The mul- pound was added to the cultures at a final concentration of5 mM tiple band pattern of HMG-14 and -17 from butyrate-treated cells about 20 hr prior to harvesting of the cells. results from hyperphosphorylation rather than hyperacetylation. Preparation of Nuclei. HeLa cell nuclei were prepared as Phosphorylation of these two small nonhistone proteins may play described by Milcarek et al. -

Anti-Dicer (SAB4200087)
Anti-Dicer produced in rabbit, affinity isolated antibody Product Number SAB4200087 Product Description Precautions and Disclaimer Anti-Dicer is produced in rabbit using as the For R&D use only. Not for drug, household, or other immunogen a synthetic peptide corresponding to a uses. Please consult the Safety Data Sheet for fragment of human Dicer (Gene ID: 23405) conjugated information regarding hazards and safe handling to KLH. The corresponding sequence is identical in practices. mouse. The antibody is affinity-purified using the immunizing peptide immobilized on agarose. Storage/Stability Store at –20 C. For continuous use, store at 2–8 C for Anti-Dicer recognizes human Dicer. The antibody may up to one month. For extended storage, freeze in be used in several immunochemical techniques working aliquots at –20 C. Repeated freezing and including immunoblotting (218 kDa), immuno- thawing, or storage in “frost-free” freezers, is not precipitation, and immunofluorescence. Detection of recommended. If slight turbidity occurs upon prolonged the Dicer band by immunoblotting is specifically storage, clarify the solution by centrifugation before inhibited with the immunizing peptide. use. Working dilutions should be discarded if not used within 12 hours. Dicer, also known as Dicer1, Endoribonuclease Dicer, Helicase with RNase motif, and HERNA, is a member Product Profile of the RNase III family that catalyzes the first step in the Immunoblotting: a working antibody concentration of RNA interference (RNAi) pathway and initiates 3-6 g/mL is recommended using HeLa cell lysates. formation of the RNA-induced silencing complex (RISC). Dicer processes the dsRNA into small Immunoprecipitation: a working antibody amount of fragments called short interfering RNA (siRNA) or 2.5-5 g is recommended using HeLa cell lysates. -
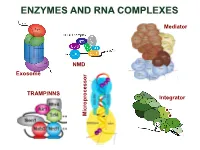
Enzymes and Rna Complexes
ENZYMES AND RNA COMPLEXES Mediator NMD Exosome NMD TRAMP/NNS Integrator Microprocessor RNA PROCESSING and DECAY machinery: RNases Protein Function Characteristics Exonucleases 5’ 3’ Xrn1 cytoplasmic, mRNA degradation processsive Rat1 nuclear, pre-rRNA, sn/snoRNA, pre-mRNA processing and degradation Rrp17/hNol12 nuclear, pre-rRNA processing Exosome 3’ 5’ multisubunit exo/endo complex subunits organized as in bacterial PNPase Rrp44/Dis3 catalytic subunit Exo/PIN domains, processsive Rrp4, Rrp40 pre-rRNA, sn/snoRNA processing, mRNA degradation Rrp41-43, 45-46 participates in NMD, ARE-dependent, non-stop decay Mtr3, Ski4 Mtr4 nuclear helicase cofactor DEAD box Rrp6 (Rrp47) nuclear exonuclease ( Rrp6 BP, cofactor) RNAse D homolog, processsive Ski2,3,7,8 cytoplasmic exosome cofactors. SKI complex helicase, GTPase Other 3’ 5’ Rex1-4 3’-5’ exonucleases, rRNA, snoRNA, tRNA processing RNase D homolog DXO 3’-5’ exonuclease in addition to decapping mtEXO 3’ 5’ mitochondrial degradosome RNA degradation in yeast Suv3/ Dss1 helicase/ 3’-5’ exonuclease DExH box/ RNase II homolog Deadenylation Ccr4/NOT/Pop2 major deadenylase complex (Ccr, Caf, Pop, Not proteins) Ccr4- Mg2+ dependent endonuclease Pan2p/Pan3 additional deadenylases (poliA tail length) RNase D homolog, poly(A) specific nuclease PARN mammalian deadenylase RNase D homolog, poly(A) specific nuclease Endonucleases RNase III -Rnt1 pre-rRNA, sn/snoRNA processing, mRNA degradation dsRNA specific -Dicer, Drosha siRNA/miRNA biogenesis, functions in RNAi PAZ, RNA BD, RNase III domains Ago2 Slicer -

A Disease-Linked Lncrna Mutation in Rnase MRP Inhibits Ribosome Synthesis
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A disease-linked lncRNA mutation in RNase MRP inhibits ribosome synthesis Nic Roberston1, Vadim Shchepachev1, David Wright2, Tomasz W. Turowski1, Christos Spanos1, Aleksandra Helwak1, Rose Zamoyska2, David Tollervey1 1 Wellcome Centre for Cell Biology, University of Edinburgh, Edinburgh, UK 2 Ashworth Laboratories, Institute of Immunology and Infection Research, University of Edinburgh, Edinburgh, UK Keywords: protein-RNA interaction; RNA-binding sites; UV crosslinking; mass spectrometry; genetic disease; Cartilage Hair Hypoplasia; ribosome synthesis; T cell activation Running title: RNase MRP defects cause ribosomopathy Highlights: • Mutations in RMRP lncRNA impair pre-rRNA processing and T cell activation • Patient derived fibroblasts show impaired pre-rRNA processing • Cells with the most common disease-linked mutation have specific processing defects • Cytoplasmic ribosomes and intact RNase MRP complexes are also reduced in these cells 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.29.437572; this version posted March 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Mutations in the human RMRP gene cause Cartilage Hair Hypoplasia (CHH), an autosomal recessive disorder characterized by skeletal abnormalities and impaired T cell activation. -

HOXB6 Homeo Box B6 HOXB5 Homeo Box B5 WNT5A Wingless-Type
5 6 6 5 . 4 2 1 1 1 2 4 6 4 3 2 9 9 7 0 5 7 5 8 6 4 0 8 2 3 1 8 3 7 1 0 0 4 0 2 5 0 8 7 5 4 1 1 0 3 6 0 4 8 3 7 4 7 6 9 6 7 1 5 0 8 1 4 1 1 7 1 0 0 4 2 0 8 1 1 1 2 5 3 5 0 7 2 6 9 1 2 1 8 3 5 2 9 8 0 6 0 9 5 1 9 9 2 1 1 6 0 2 3 0 3 6 9 1 6 5 5 7 1 1 2 1 1 7 5 4 6 6 4 1 1 2 8 4 7 1 6 2 7 7 5 4 3 2 4 3 6 9 4 1 7 1 3 4 1 2 1 3 1 1 4 7 3 1 1 1 1 5 3 2 6 1 5 1 3 5 4 5 2 3 1 1 6 1 7 3 2 5 4 3 1 6 1 5 3 1 7 6 5 1 1 1 4 6 1 6 2 7 2 1 2 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a a S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S S HOXB6 homeo box B6 HOXB5 homeo box B5 WNT5A wingless-type MMTV integration site family, member 5A WNT5A wingless-type MMTV integration site family, member 5A FKBP11 FK506 binding protein 11, 19 kDa EPOR erythropoietin receptor SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 SLC5A6 solute carrier family 5 sodium-dependent vitamin transporter, member 6 RAD52 RAD52 homolog S. -

A Specialized Processing Body That Is Temporally and Asymmetrically Regulated During the Cell Cycle in Saccharomyces Cerevisiae
JCB: ARTICLE A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae Tina Gill, Jason Aulds, and Mark E. Schmitt Department of Biochemistry and Molecular Biology, State University of New York Upstate Medical University, Syracuse, NY 13210 Nase mitochondrial RNA processing (MRP) is an this spot was asymmetrically found in daughter cells, essential ribonucleoprotein endoribonuclease that where the RNase MRP substrate, CLB2 mRNA, localizes. R functions in the degradation of specifi c mRNAs in- Both the mitotic exit network and fourteen early ana- volved in cell cycle regulation. We have investigated phase release pathways are nonessential but important where this processing event occurs and how it is regu- for the temporal changes in localization. Asymmetric lo- lated. As expected, results demonstrate that RNase MRP calization was found to be dependent on the locasome. is predominantly localized in the nucleolus, where it The evidence suggests that these spots are specialized processes ribosomal RNAs. However, after the initiation processing bodies for the degradation of transcripts that of mitosis, RNase MRP localizes throughout the entire are cell cycle regulated and daughter cell localized. We nucleus and in a single discrete cytoplasmic spot that have called these TAM bodies for temporal asymmetric persists until the completion of telophase. Furthermore, MRP bodies. Introduction RNase mitochondrial RNA processing (MRP) is an essential ri- 27SA preribosomal RNA at the A3 site, forming the 5.8S(s) bonucleoprotein endoribonuclease that cleaves RNA substrates ribosomal RNA (rRNA; Schmitt and Clayton, 1993; Lygerou in a site-specifi c manner and is highly conserved in eukaryotes et al., 1996). -

Datasheet for T7 Exonuclease (M0263; Lot 0031212)
Source: Purified from an E. coli strain containing a Unit Assay Conditions: 50 mM potassium Physical Purity: Purified to > 95% homogeneity TYB12 intein fusion acetate, 20 mM Tris-acetate, 10 mM magnesium as determined by SDS-PAGE analysis using T7 Exonuclease acetate, 1mM dithiothreitol (pH 7.9) and 0.15 mM Coomassie Blue detection. Supplied in: 10 mM Tris-HCl (pH 8.0), sonicated duplex [3H] DNA. 0.1 mM EDTA, 5 mM DTT and 50% glycerol. RNase Activity (Extended Digestion): A 10 µl Quality Control Assays 1-800-632-7799 reaction in NEBuffer 4 containing 40 ng of [email protected] Reagents Supplied with Enzyme: Single Stranded Deoxyribonuclease Activity flourescein labeled RNA transcript and 10 units www.neb.com 10X NEBuffer 4. (FAM Labeled Oligo): A 50 µl reaction in of T7 Exonuclease incubated at 37°C. After M0263S 003121214121 NEBuffer 4 containing a 20 nM solution of a incubation for 4 hours, > 90% of the substrate Reaction Conditions: fluorescent internal labeled oligonucleotide and a RNA remains intact as determined by gel minimum of 50 units of T7 Exonuclease incubated 1X NEBuffer 4. electrophoresis using fluorescence detection. M0263S Incubate at 25°C. for 16 hours at 37°C yields < 5% degradation as determined by capillary electrophoresis. 1,000 units 10,000 U/ml Lot: 0031212 Heat Inactivation: No 1X NEBuffer 4: RECOMBINANT Store at –20°C Exp: 12/14 50 mM potassium acetate Endonuclease Activity: Incubation of a 50 µl References: reaction containing 100 units of T7 Exonuclease Description: T7 Exonuclease acts in the 5´ to 20 mM Tris-acetate 1. -

Termin Translat Trna Utr Mutat Protein Signal
Drugs & Chemicals 1: Tumor Suppressor Protein p53 2: Heterogeneous-Nuclear Ribonucleo- (1029) proteins (14) activ apoptosi arf cell express function inactiv induc altern assai associ bind mdm2 mutat p53 p73 pathwai protein regul complex detect exon famili genom respons suppress suppressor tumor wild-typ interact intron isoform nuclear protein sensit site specif splice suggest variant 3: RNA, Transfer (110) 4: DNA Primers (1987) codon contain differ eukaryot gene initi amplifi analysi chain clone detect dna express mrna protein region ribosom rna fragment gene genotyp mutat pcr sequenc site speci suggest synthesi polymorph popul primer reaction region restrict sequenc speci termin translat trna utr 5: Saccharomyces cerevisiae Proteins 6: Apoptosis Regulatory Proteins (291) (733) activ apoptosi apoptosis-induc albican bud candida cerevisia complex encod apoptot bcl-2 caspas caspase-8 cell eukaryot fission function growth interact involv death fasl induc induct ligand methyl necrosi pathwai program sensit surviv trail mutant pomb protein requir saccharomyc strain suggest yeast 7: Plant Proteins (414) 8: Membrane Proteins (1608) access arabidopsi cultivar flower hybrid leaf leav apoptosi cell conserv domain express function gene human identifi inhibitor line maiz plant pollen rice root seed mammalian membran mice mous mutant seedl speci thaliana tomato transgen wheat mutat protein signal suggest transport 1 9: Tumor Suppressor Proteins (815) 10: 1-Phosphatidylinositol 3-Kinase activ arrest cell cycl cyclin damag delet dna (441) 3-kinas activ