First Phylogeny of Bitterbush Family, Picramniaceae (Picramniales)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

"National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary."
Intro 1996 National List of Vascular Plant Species That Occur in Wetlands The Fish and Wildlife Service has prepared a National List of Vascular Plant Species That Occur in Wetlands: 1996 National Summary (1996 National List). The 1996 National List is a draft revision of the National List of Plant Species That Occur in Wetlands: 1988 National Summary (Reed 1988) (1988 National List). The 1996 National List is provided to encourage additional public review and comments on the draft regional wetland indicator assignments. The 1996 National List reflects a significant amount of new information that has become available since 1988 on the wetland affinity of vascular plants. This new information has resulted from the extensive use of the 1988 National List in the field by individuals involved in wetland and other resource inventories, wetland identification and delineation, and wetland research. Interim Regional Interagency Review Panel (Regional Panel) changes in indicator status as well as additions and deletions to the 1988 National List were documented in Regional supplements. The National List was originally developed as an appendix to the Classification of Wetlands and Deepwater Habitats of the United States (Cowardin et al.1979) to aid in the consistent application of this classification system for wetlands in the field.. The 1996 National List also was developed to aid in determining the presence of hydrophytic vegetation in the Clean Water Act Section 404 wetland regulatory program and in the implementation of the swampbuster provisions of the Food Security Act. While not required by law or regulation, the Fish and Wildlife Service is making the 1996 National List available for review and comment. -

Redalyc.Tripanocide and Antibacterial Activity of Alvaradoa Subovata Cronquist Extracts
Boletín Latinoamericano y del Caribe de Plantas Medicinales y Aromáticas ISSN: 0717-7917 [email protected] Universidad de Santiago de Chile Chile MARTÍNEZ, M. Laura; TRAVAINI, M. Lucía; RODRIGUEZ, M. Victoria; ORELLANO, Elena; NOCITO, Isabel; SERRA, Esteban; GATTUSO, Martha; CORTADI, Adriana Tripanocide and antibacterial activity of Alvaradoa subovata Cronquist extracts Boletín Latinoamericano y del Caribe de Plantas Medicinales y Aromáticas, vol. 12, núm. 3, mayo, 2013, pp. 302-312 Universidad de Santiago de Chile Santiago, Chile Available in: http://www.redalyc.org/articulo.oa?id=85626383006 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative © 2013 Boletín Latinoamericano y del Caribe de Plantas Medicinales y Aromáticas 12 (3): 302 - 312 ISSN 0717 7917 www.blacpma.usach.cl Artículo Original | Original Article Tripanocide and antibacterial activity of Alvaradoa subovata Cronquist extracts [Actividad tripanocida y antibacteriana de extractos de Alvaradoa subovata Cronquist] M. Laura MARTÍNEZ1, M. Lucía TRAVAINI1, M. Victoria RODRIGUEZ1, Elena ORELLANO2, Isabel NOCITO3, Esteban SERRA3, Martha GATTUSO1 & Adriana CORTADI1. 1Área Biología Vegetal 2Área Biología Molecular 3Área Parasitología. Facultad de Ciencias Bioquímicas y Farmacéuticas Universidad Nacional de Rosario. Suipacha 531. S2002LRK Rosario, Argentina Contactos | Contacts: M. Laura MARTÍNEZ - E-mail address: [email protected] Abstract We studied antioxidant, antibacterial and tripanocide activities of Alvaradoa subovata extracts. The ethanolic extracts showed the greatest DPPH radical scavenging capacity, especially that of bark with an IC50 = 4.7 ± 0.18 µg/mL. -

Low Risk, Fruit Tree, Edible Fruit, Slow-Growing, Bird-Dispersed, Zoochorous
Family: Sapindaceae Taxon: Talisia esculenta Synonym: Sapindus esculenta A. St.-Hil. (basionym) Common Name: pitomba Questionaire : current 20090513 Assessor: Chuck Chimera Designation: L Status: Assessor Approved Data Entry Person: Chuck Chimera WRA Score -1 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? y=1, n=-1 103 Does the species have weedy races? y=1, n=-1 201 Species suited to tropical or subtropical climate(s) - If island is primarily wet habitat, then (0-low; 1-intermediate; 2- High substitute "wet tropical" for "tropical or subtropical" high) (See Appendix 2) 202 Quality of climate match data (0-low; 1-intermediate; 2- High high) (See Appendix 2) 203 Broad climate suitability (environmental versatility) y=1, n=0 204 Native or naturalized in regions with tropical or subtropical climates y=1, n=0 y 205 Does the species have a history of repeated introductions outside its natural range? y=-2, ?=-1, n=0 n 301 Naturalized beyond native range y = 1*multiplier (see n Appendix 2), n= question 205 302 Garden/amenity/disturbance weed n=0, y = 1*multiplier (see n Appendix 2) 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see n Appendix 2) 304 Environmental weed n=0, y = 2*multiplier (see n Appendix 2) 305 Congeneric weed n=0, y = 1*multiplier (see n Appendix 2) 401 Produces spines, thorns or burrs y=1, n=0 n 402 Allelopathic y=1, n=0 403 Parasitic y=1, n=0 n 404 Unpalatable to grazing animals y=1, n=-1 405 Toxic to animals y=1, n=0 406 Host for recognized pests -

Well-Known Plants in Each Angiosperm Order
Well-known plants in each angiosperm order This list is generally from least evolved (most ancient) to most evolved (most modern). (I’m not sure if this applies for Eudicots; I’m listing them in the same order as APG II.) The first few plants are mostly primitive pond and aquarium plants. Next is Illicium (anise tree) from Austrobaileyales, then the magnoliids (Canellales thru Piperales), then monocots (Acorales through Zingiberales), and finally eudicots (Buxales through Dipsacales). The plants before the eudicots in this list are considered basal angiosperms. This list focuses only on angiosperms and does not look at earlier plants such as mosses, ferns, and conifers. Basal angiosperms – mostly aquatic plants Unplaced in order, placed in Amborellaceae family • Amborella trichopoda – one of the most ancient flowering plants Unplaced in order, placed in Nymphaeaceae family • Water lily • Cabomba (fanwort) • Brasenia (watershield) Ceratophyllales • Hornwort Austrobaileyales • Illicium (anise tree, star anise) Basal angiosperms - magnoliids Canellales • Drimys (winter's bark) • Tasmanian pepper Laurales • Bay laurel • Cinnamon • Avocado • Sassafras • Camphor tree • Calycanthus (sweetshrub, spicebush) • Lindera (spicebush, Benjamin bush) Magnoliales • Custard-apple • Pawpaw • guanábana (soursop) • Sugar-apple or sweetsop • Cherimoya • Magnolia • Tuliptree • Michelia • Nutmeg • Clove Piperales • Black pepper • Kava • Lizard’s tail • Aristolochia (birthwort, pipevine, Dutchman's pipe) • Asarum (wild ginger) Basal angiosperms - monocots Acorales -

Ethnopharmacology of Fruit Plants
molecules Review Ethnopharmacology of Fruit Plants: A Literature Review on the Toxicological, Phytochemical, Cultural Aspects, and a Mechanistic Approach to the Pharmacological Effects of Four Widely Used Species Aline T. de Carvalho 1, Marina M. Paes 1 , Mila S. Cunha 1, Gustavo C. Brandão 2, Ana M. Mapeli 3 , Vanessa C. Rescia 1 , Silvia A. Oesterreich 4 and Gustavo R. Villas-Boas 1,* 1 Research Group on Development of Pharmaceutical Products (P&DProFar), Center for Biological and Health Sciences, Federal University of Western Bahia, Rua Bertioga, 892, Morada Nobre II, Barreiras-BA CEP 47810-059, Brazil; [email protected] (A.T.d.C.); [email protected] (M.M.P.); [email protected] (M.S.C.); [email protected] (V.C.R.) 2 Physical Education Course, Center for Health Studies and Research (NEPSAU), Univel University Center, Cascavel-PR, Av. Tito Muffato, 2317, Santa Cruz, Cascavel-PR CEP 85806-080, Brazil; [email protected] 3 Research Group on Biomolecules and Catalyze, Center for Biological and Health Sciences, Federal University of Western Bahia, Rua Bertioga, 892, Morada Nobre II, Barreiras-BA CEP 47810-059, Brazil; [email protected] 4 Faculty of Health Sciences, Federal University of Grande Dourados, Dourados, Rodovia Dourados, Itahum Km 12, Cidade Universitaria, Caixa. postal 364, Dourados-MS CEP 79804-970, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-(77)-3614-3152 Academic Editors: Raffaele Pezzani and Sara Vitalini Received: 22 July 2020; Accepted: 31 July 2020; Published: 26 August 2020 Abstract: Fruit plants have been widely used by the population as a source of food, income and in the treatment of various diseases due to their nutritional and pharmacological properties. -

Alphabetical Lists of the Vascular Plant Families with Their Phylogenetic
Colligo 2 (1) : 3-10 BOTANIQUE Alphabetical lists of the vascular plant families with their phylogenetic classification numbers Listes alphabétiques des familles de plantes vasculaires avec leurs numéros de classement phylogénétique FRÉDÉRIC DANET* *Mairie de Lyon, Espaces verts, Jardin botanique, Herbier, 69205 Lyon cedex 01, France - [email protected] Citation : Danet F., 2019. Alphabetical lists of the vascular plant families with their phylogenetic classification numbers. Colligo, 2(1) : 3- 10. https://perma.cc/2WFD-A2A7 KEY-WORDS Angiosperms family arrangement Summary: This paper provides, for herbarium cura- Gymnosperms Classification tors, the alphabetical lists of the recognized families Pteridophytes APG system in pteridophytes, gymnosperms and angiosperms Ferns PPG system with their phylogenetic classification numbers. Lycophytes phylogeny Herbarium MOTS-CLÉS Angiospermes rangement des familles Résumé : Cet article produit, pour les conservateurs Gymnospermes Classification d’herbier, les listes alphabétiques des familles recon- Ptéridophytes système APG nues pour les ptéridophytes, les gymnospermes et Fougères système PPG les angiospermes avec leurs numéros de classement Lycophytes phylogénie phylogénétique. Herbier Introduction These alphabetical lists have been established for the systems of A.-L de Jussieu, A.-P. de Can- The organization of herbarium collections con- dolle, Bentham & Hooker, etc. that are still used sists in arranging the specimens logically to in the management of historical herbaria find and reclassify them easily in the appro- whose original classification is voluntarily pre- priate storage units. In the vascular plant col- served. lections, commonly used methods are systema- Recent classification systems based on molecu- tic classification, alphabetical classification, or lar phylogenies have developed, and herbaria combinations of both. -

Patterns of Plant Diversity and Endemism in Namibia
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Stellenbosch University SUNScholar Repository Bothalia 36,2: 175-189(2006) Patterns of plant diversity and endemism in Namibia P. CRAVEN* and P VORSTER** Keywords: Namibia, phytogeography, plant endemism ABSTRACT Species richness, endemism and areas that are rich in both species and endemic species were assessed and mapped for Namibia. High species diversity corresponds with zones where species overlap. These are particularly obvious where there are altitudinal variations and in high-lying areas. The endemic flora o f Namibia is rich and diverse. An estimated 16% of the total plant species in Namibia are endemic to the country. Endemics are in a wide variety o f families and sixteen genera are endemic. Factors that increase the likelihood o f endemism are mountains, hot deserts, diversity o f substrates and microclimates. The distribution of plants endemic to Namibia was arranged in three different ways. Firstly, based on a grid count with the phytogeographic value of the species being equal, overall endemism was mapped. Secondly, range restricted plant species were mapped individually and those with congruent distribution patterns were combined. Thirdly, localities that are important for very range-restricted species were identified. The resulting maps of endemism and diversity were compared and found to correspond in many localities. When overall endemism is compared with overall diversity, rich localities may consist o f endemic species with wide ranges. The other methods identify important localities with their own distinctive complement of species. INTRODUCTION (1994). It was based on distributional data per magiste rial district following Merxmiiller (1966-1972), as well Species diversity was traditionally measured by count as other literature. -

Kirkia Wilmsii Engl
A Maroyi /J. Pharm. Sci. & Res. Vol. 11(7), 2019, 2628-2631 Kirkia wilmsii Engl.: A review of its botany, medicinal uses, phytochemistry and biological activities A Maroyi Medicinal Plants and Economic Development (MPED) Research Centre, Department of Botany, University of Fort Hare, Private Bag X1314, Alice 5700, South Africa Abstract Kirkia wilmsii is a small-sized deciduous tree widely used as herbal medicine in South Africa. This study is aimed at providing a critical review of the botany, biological activities, phytochemistry and medicinal uses of K. wilmsii. Documented information on the botany, biological activities, medicinal uses and phytochemistry of K. wilmsii was collected from several online sources which included BMC, Scopus, SciFinder, Google Scholar, Science Direct, Elsevier, Pubmed and Web of Science. Additional information on the botany, biological activities, phytochemistry and medicinal uses of K. wilmsii was gathered from pre-electronic sources such as book chapters, books, journal articles and scientific publications sourced from the University library. This study showed that the bark, leaves, rootbark and tubers of K. wilmsii are used as herbal medicine for arthritis, asthma, diabetes mellitus, diarrhoea, high blood pressure, hypertension, malaria, nasal congestion, respiratory infections, ringworm and tuberculosis. Phytochemical analyses revealed that the leaves, tubers and twigs of K. wilmsii are characterized by caffeic acid, cardenolide deoxy sugars, cardiac glycosides, ellagic acid, flavonoids, gallic acid, isocoumarin, neo-lignan, nor-carotenoids, phenolics, phlobatannins, quercetin, reducing sugars, saponins, steroids, tannins and terpenoids. Pharmacological research revealed that K. wilmsii crude extracts have antimicrobial, antioxidant, antiplasmodial, antiplatelet and cytotoxicity activities. Future ethnopharmacological research should focus on correlating ethnomedicinal uses of the species with its pharmacological properties. -

Illinois Native Plant Society 2019 Plant List Herbaceous Plants
Illinois Native Plant Society 2019 Plant List Plant Sale: Saturday, May 11, 9:00am – 1:00pm Illinois State Fairgrounds Commodity Pavilion (Across from Grandstand) Herbaceous Plants Scientific Name Common Name Description Growing Conditions Comment Dry to moist, Sun to part Blooms mid-late summer Butterfly, bee. Agastache foeniculum Anise Hyssop 2-4', Lavender to purple flowers shade AKA Blue Giant Hyssop Allium cernuum var. Moist to dry, Sun to part Nodding Onion 12-18", Showy white flowers Blooms mid summer Bee. Mammals avoid cernuum shade 18", White flowers after leaves die Allium tricoccum Ramp (Wild Leek) Moist, Shade Blooms summer Bee. Edible back Moist to dry, Part shade to Blooms late spring-early summer Aquilegia canadensis Red Columbine 30", Scarlet and yellow flowers shade Hummingbird, bee Moist to wet, Part to full Blooms late spring-early summer Showy red Arisaema dracontium Green Dragon 1-3', Narrow greenish spadix shade fruits. Mammals avoid 1-2', Green-purple spadix, striped Moist to wet, Part to full Blooms mid-late spring Showy red fruits. Arisaema triphyllum Jack-in-the-Pulpit inside shade Mammals avoid Wet to moist, Part shade to Blooms late spring-early summer Bee. AKA Aruncus dioicus Goatsbeard 2-4', White fluffy panicles in spring shade Brides Feathers Wet to moist, Light to full Blooms mid-late spring Mammals avoid. Asarum canadense Canadian Wildginger 6-12", Purplish brown flowers shade Attractive groundcover 3-5', White flowers with Moist, Dappled sun to part Blooms summer Monarch larval food. Bee, Asclepias exaltata Poke Milkweed purple/green tint shade butterfly. Uncommon Blooms mid-late summer Monarch larval Asclepias hirtella (Tall) Green Milkweed To 3', Showy white flowers Dry to moist, Sun food. -

Alvaradoa Amorphoides Germination at Low Water Potential and the Role of the Antioxidant System
Botanical Sciences 93 (2): -9, 205 PHYSIOLOGY DOI: 0.729/botsci.6 ALVARADOA AMORPHOIDES GERMINATION AT LOW WATER POTENTIAL AND THE ROLE OF THE ANTIOXIDANT SYSTEM VERÓNICA HERNÁNDEZ-PÉREZ1, JUDITH MÁRQUEZ-GUZMÁN2, SOBEIDA SÁNCHEZ-NIETO3 1, 4 AND ROCÍO CRUZ-ORTEGA 1Instituto de Ecología, Departamento de Ecología Funcional, Universidad Nacional Autónoma de México, México, D.F., Mexico 2Facultad de Ciencias, Laboratorio de Desarrollo de Plantas, Universidad Nacional Autónoma de México, México, D.F., Mexico 3Facultad de Química, Departamento de Bioquímica, Universidad Nacional Autónoma de México, México, D.F., Mexico 4Author for correspondence: Rocío Cruz-Ortega: [email protected] Abstract: Tropical dry forests are characterized by a high diversity of tree communities and extremely heterogeneous water avai- lability. The tree Alvaradoa amorphoides is a pioneer species of the tropical dry forest found in Xochicalco, Morelos, Mexico. To determine the water requirements for this species to germinate, we evaluated the seed germination rates under field and labo- ratory conditions. In the field, the seeds had an overall mean germination rate of 42%, but the rate varied between the different sites independent of the soil relative humidity and landscape. Alvaradoa amorphoides seeds exposed to a water potential of -0.5 MPa delayed germination, extending Phase II. At the -.0 and -.5 MPa water potentials, germination was inhibited by 80 and 00%, respectively, but the seeds remained viable. Although, the oxygen consumption did not differ between the treatments, the respiration profiles did not show the same triphasic curve as the control. The H2O2 and O2- levels were not significantly different in the seeds at the evaluated low-water potentials (-0.5 and -.0 MPa), nor were the catalase, superoxide dismutase, and gluta- thione reductase activity. -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M. -
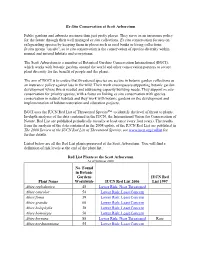
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details.