Hidden Complexity of Lichen Symbiosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of Genetic Systems in Filamentous Ascomycetes
Evolution of Genetic Systems in Filamentous Ascomycetes Evolutie van genetische systemen in hyphenvormende zakjeszwammen 0000 0513 3836 Promotor: dr. R.F. Hoekstra hoogleraar in de populatie- en kwantitatieve genetica fjtfoiißi f ßin Maarten J. Nauta Evolution of Genetic Systems in Filamentous Ascomycetes Proefschrift ter verkrijging van de graad van doctor in de landbouw- en milieuwetenschappen op gezag van de rector magnificus, dr. C.M. Karssen, in het openbaar te verdedigen op woensdag 12januar i 1994 des namiddags te vier uur in de Aula van de Landbouwuniversiteit te Wageningen. 15 0 S(p^ZJ> These investigations were supported by the Netherlands Organization for Scientific Research (N.W.O.). BibUt/FHEEK LAMDbOirWUNIVERSITEJi. WAGE NINGE N CIP-GEGEVENS KONINKLIJKE BIBLIOTHEEK, DEN HAAG Nauta, Maarten J. Evolution of genetic systems in filamentous ascomycetes / Maarten J. Nauta. - [ S.l. : s.n.]. -111 . Thesis Wageningen. - With ref. - With summary in Dutch. ISBN 90-5485-199-6 Subject headings: population genetics / ascomycetes. omslagontwerp: Ernst van Cleef foto omslag: Barrages tussen verschillende stammen van Podospora anserina als gevolg van vegetatieve incompatibiliteit. (met dank aan Inge Haspels) aan mijn ouders Voorwoord Dit proefschrift is het resultaat van vier jaar onderzoek, verricht bij de vakgroep Erfelijkheidsleer van de Landbouwuniversiteit in Wageningen. In zekere zin valt zo'n proefschrift te vergelijken met een levend wezen. Uit de genetica is bekend dat de verschijningsvorm van elk levend wezen tot stand komt door een combinatie van erfelijke aanleg en invloeden uit de omgeving. Voor een proefschrift geldt eigenlijk hetzelfde: Zowel het werk van de auteur, als de bijdragen van zijn omgeving zijn onontbeerlijk om tot een verschijningsvorm te komen. -

The Lichens' Microbiota, Still a Mystery?
fmicb-12-623839 March 24, 2021 Time: 15:25 # 1 REVIEW published: 30 March 2021 doi: 10.3389/fmicb.2021.623839 The Lichens’ Microbiota, Still a Mystery? Maria Grimm1*, Martin Grube2, Ulf Schiefelbein3, Daniela Zühlke1, Jörg Bernhardt1 and Katharina Riedel1 1 Institute of Microbiology, University Greifswald, Greifswald, Germany, 2 Institute of Plant Sciences, Karl-Franzens-University Graz, Graz, Austria, 3 Botanical Garden, University of Rostock, Rostock, Germany Lichens represent self-supporting symbioses, which occur in a wide range of terrestrial habitats and which contribute significantly to mineral cycling and energy flow at a global scale. Lichens usually grow much slower than higher plants. Nevertheless, lichens can contribute substantially to biomass production. This review focuses on the lichen symbiosis in general and especially on the model species Lobaria pulmonaria L. Hoffm., which is a large foliose lichen that occurs worldwide on tree trunks in undisturbed forests with long ecological continuity. In comparison to many other lichens, L. pulmonaria is less tolerant to desiccation and highly sensitive to air pollution. The name- giving mycobiont (belonging to the Ascomycota), provides a protective layer covering a layer of the green-algal photobiont (Dictyochloropsis reticulata) and interspersed cyanobacterial cell clusters (Nostoc spec.). Recently performed metaproteome analyses Edited by: confirm the partition of functions in lichen partnerships. The ample functional diversity Nathalie Connil, Université de Rouen, France of the mycobiont contrasts the predominant function of the photobiont in production Reviewed by: (and secretion) of energy-rich carbohydrates, and the cyanobiont’s contribution by Dirk Benndorf, nitrogen fixation. In addition, high throughput and state-of-the-art metagenomics and Otto von Guericke University community fingerprinting, metatranscriptomics, and MS-based metaproteomics identify Magdeburg, Germany Guilherme Lanzi Sassaki, the bacterial community present on L. -

North American Fungi
North American Fungi Volume 3, Number 4, Pages 1-15 Published April 27, 2008 Formerly Pacific Northwest Fungi Changes in forage lichen biomass after insect outbreaks and fuel reduction treatments in the Blue Mountains, Oregon Bruce McCune, Sarah Jovan and Amanda Hardman Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR 97331-2902 McCune, B., S. Jovan and A. Hardman. 2008. Changes in forage lichen biomass after insect outbreaks and fuel reduction treatments in the Blue Mountains, Oregon. North American Fungi 3(4): 1-15. doi: 10.2509/naf2008.003.00a Corresponding author: Bruce McCune, [email protected]. Accepted for publication April 19, 2008. http://pnwfungi.org Copyright © 2008 Pacific Northwest Fungi Project. All rights reserved. Abstract: Forage lichens are pendulous, hairlike species eaten by a wide range of mammals. Our overall goal was to estimate losses of Bryoria, a genus of ecologically important forage species, in forests subjected to disease and fuel reduction treatments at Starkey Experimental Forest in the Blue Mountains of northeastern Oregon. Specific objectives were to (1) estimate Bryoria biomass in stands decimated by insects and disease, (2) compare Bryoria biomass in untreated stands with those treated by mechanical fuels reduction and prescribed fire, and (3) estimate the range of pre-insect outbreak Bryoria biomass using historical data. Our general approach was to estimate tree-level Bryoria biomass on a sample of trees, regress estimates against tree size and species using nonparametric multiplicative regression (NPMR), then predict stand-level biomass by applying NPMR to tree size and density data. For live trees, logarithm of dbh was a strong predictor of Bryoria biomass (cross validated R2 = xR2 = 0.83). -

Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho
Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho To cite this version: James Umen, Susana Coelho. Algal Sex Determination and the Evolution of Anisogamy. Annual Review of Microbiology, Annual Reviews, 2019, 73 (1), 10.1146/annurev-micro-020518-120011. hal- 02187088 HAL Id: hal-02187088 https://hal.sorbonne-universite.fr/hal-02187088 Submitted on 17 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Annu. Rev. Microbiol. 2019. 73:X–X https://doi.org/10.1146/annurev-micro-020518-120011 Copyright © 2019 by Annual Reviews. All rights reserved Umen • Coelho www.annualreviews.org • Algal Sexes and Mating Systems Algal Sex Determination and the Evolution of Anisogamy James Umen1 and Susana Coelho2 1Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA; email: [email protected] 2Sorbonne Université, UPMC Université Paris 06, CNRS, Algal Genetics Group, UMR 8227, Integrative Biology of Marine Models, Station Biologique de Roscoff, CS 90074, F-29688, Roscoff, France [**AU: Please write the entire affiliation in French or write it all in English, rather than a combination of English and French**] ; email: [email protected] Abstract Algae are photosynthetic eukaryotes whose taxonomic breadth covers a range of life histories, degrees of cellular and developmental complexity, and diverse patterns of sexual reproduction. -

Morphological Traits in Hair Lichens Affect Their Water Storage
Morphological traits in hair lichens affect their water storage Therese Olsson Student Degree Thesis in Biology 30 ECTS Master’s Level Report passed: 29 August 2014 Supervisor: Per-Anders Esseen Abstract The aim with this study was to develop a method to estimate total area of hair lichens and to compare morphological traits and water storage in them. Hair lichens are an important component of the epiphytic flora in boreal forests. Their growth is primarily regulated by available water, and light when hydrated. Lichens have no active mechanism to regulate their 2 water content and their water holding capacity (WHC, mg H2O/cm ) is thus an important factor for how long they remain wet and metabolically active. In this study, the water uptake and loss in five hair lichens (Alectoria sarmentosa, three Bryoria spp. and Usnea dasypoga) were compared. Their area were estimated by combining photography, scanning and a computer programme that estimates the area of objects. Total area overlap of individual branches was calculated for each species, to estimate total area of the lichen. WHC and specific thallus mass (STM) (mg DM/cm2) of the lichens were calculated. Bryoria spp. had a significantly lower STM compared to U. dasypoga and A. sarmentosa, due to its thinner branches and higher branch density. Bryoria also had a lower WHC compared to A. sarmentosa, promoting a rapid uptake and loss of water. All species had a significant relationship between STM and WHC, above a 1:1 line for all species except U. dasypoga. The lower relationship in U. dasypoga is explained by its less developed branching in combination with its thick branches. -

Sex in the Extremes: Lichen-Forming Fungi
Mycologist, Volume 19, Part 2 May 2005. ©Cambridge University Press Printed in the United Kingdom. DOI: 10.1017/S0269915XO5002016 Sex in the extremes: lichen-forming fungi FABIAN A. SEYMOUR, PETER D. CRITTENDEN & PAUL S. DYER* School of Biology, University of Nottingham, University Park, Nottingham, NG7 2RD, UK. Tel. +44 (0) 115 9513203, Fax +44 (0) 115 9513251 E-mail: [email protected]; [email protected] *Corresponding Author Lichens are characteristically found in environments subject to extremes of temperature, desiccation and low nutrient status. Despite this sexual structures are often formed in abundance. The underlying mechanisms of sex in lichen-forming fungi are discussed, together with possible ecological reasons for the persistence of sexuality. Special features of lichen sex are highlighted including sex at the limits of life on earth in Antarctica, re-licheniza- tion following sex and dispersal, and the perennial nature of lichen fruiting bodies. Keywords: lichen, fungi, sex, breeding system, (98%) belonging to the Ascomycotina (Kirk et al., symbiosis, extreme environments, Antarctica 2001). They display a variety of morphologies, from flattened crust (crustose) or leafy (foliose) forms to Lichens - living together in a long-term relation- shrubby or pendulous fruticose types (Honegger, 2001) ship (Figs 3, 4, 7, 8). Lichens are seen as a textbook example of a successful Life in extreme environments mutualistic symbiosis. They consist of at least two A key characteristic of lichens is that they have a organisms: a fungus (the ‘mycobiont’), and an remarkable ability to tolerate extreme environmental intimately associated photosynthetic partner (the conditions and sustain growth despite frequent cycles ‘photobiont’). -

Interface Between Fungi and Green Algae in Lichen Associations
Botany Interface between fungi and green algae in lichen associations Journal: Botany Manuscript ID cjb-2017-0037.R2 Manuscript Type: Review Date Submitted by the Author: 07-May-2017 Complete List of Authors: Piercey-Normore, Michele; Memorial University of NL, Science and the Environment Athukorala,Draft Sarangi; University of Peradeniya, Department of Botany Is the invited manuscript for consideration in a Special Fungi on the Edge Issue? : Keyword: interaction, lichen fungi, resynthesis, symbiont communication https://mc06.manuscriptcentral.com/botany-pubs Page 1 of 34 Botany 1 Mini review - Botany Interface between fungi and green algae in lichen associations Michele D. Piercey-Normore 1 and Sarangi N. P. Athukorala 2 1School of Science and the Environment, Memorial University of NL (Grenfell campus), Corner Brook, NL, Canada, A2K 5G4, [email protected] 2Department of Botany, Faculty of Science, University of Peradeniya, Peradeniya, Sri Lanka, 20400, [email protected] Draft Corresponding author: M. D. Piercey-Normore, School of Science and the Environment, Memorial University of NL (Grenfell campus), Corner Brook, NL, Canada, A2K 5G4, email: mpiercey- [email protected] , phone: (709) 637-7166. https://mc06.manuscriptcentral.com/botany-pubs Botany Page 2 of 34 2 Abstract It is widely recognized that the lichen is the product of a fungus and a photosynthetic partner (green alga or cyanobacterium) but its acceptance was slow to develop throughout history. The development of powerful microscopic and other lab techniques enabled better understanding of the interface between symbionts beginning with the contentious concept of the dual nature of the lichen thallus. Even with accelerating progress in understanding the interface between symbionts, much more work is needed to reach a level of knowledge consistent with that of other fungal interactions. -

Plant Life MagillS Encyclopedia of Science
MAGILLS ENCYCLOPEDIA OF SCIENCE PLANT LIFE MAGILLS ENCYCLOPEDIA OF SCIENCE PLANT LIFE Volume 4 Sustainable Forestry–Zygomycetes Indexes Editor Bryan D. Ness, Ph.D. Pacific Union College, Department of Biology Project Editor Christina J. Moose Salem Press, Inc. Pasadena, California Hackensack, New Jersey Editor in Chief: Dawn P. Dawson Managing Editor: Christina J. Moose Photograph Editor: Philip Bader Manuscript Editor: Elizabeth Ferry Slocum Production Editor: Joyce I. Buchea Assistant Editor: Andrea E. Miller Page Design and Graphics: James Hutson Research Supervisor: Jeffry Jensen Layout: William Zimmerman Acquisitions Editor: Mark Rehn Illustrator: Kimberly L. Dawson Kurnizki Copyright © 2003, by Salem Press, Inc. All rights in this book are reserved. No part of this work may be used or reproduced in any manner what- soever or transmitted in any form or by any means, electronic or mechanical, including photocopy,recording, or any information storage and retrieval system, without written permission from the copyright owner except in the case of brief quotations embodied in critical articles and reviews. For information address the publisher, Salem Press, Inc., P.O. Box 50062, Pasadena, California 91115. Some of the updated and revised essays in this work originally appeared in Magill’s Survey of Science: Life Science (1991), Magill’s Survey of Science: Life Science, Supplement (1998), Natural Resources (1998), Encyclopedia of Genetics (1999), Encyclopedia of Environmental Issues (2000), World Geography (2001), and Earth Science (2001). ∞ The paper used in these volumes conforms to the American National Standard for Permanence of Paper for Printed Library Materials, Z39.48-1992 (R1997). Library of Congress Cataloging-in-Publication Data Magill’s encyclopedia of science : plant life / edited by Bryan D. -

Lichen Functional Trait Variation Along an East-West Climatic Gradient in Oregon and Among Habitats in Katmai National Park, Alaska
AN ABSTRACT OF THE THESIS OF Kaleigh Spickerman for the degree of Master of Science in Botany and Plant Pathology presented on June 11, 2015 Title: Lichen Functional Trait Variation Along an East-West Climatic Gradient in Oregon and Among Habitats in Katmai National Park, Alaska Abstract approved: ______________________________________________________ Bruce McCune Functional traits of vascular plants have been an important component of ecological studies for a number of years; however, in more recent times vascular plant ecologists have begun to formalize a set of key traits and universal system of trait measurement. Many recent studies hypothesize global generality of trait patterns, which would allow for comparison among ecosystems and biomes and provide a foundation for general rules and theories, the so-called “Holy Grail” of ecology. However, the majority of these studies focus on functional trait patterns of vascular plants, with a minority examining the patterns of cryptograms such as lichens. Lichens are an important component of many ecosystems due to their contributions to biodiversity and their key ecosystem services, such as contributions to mineral and hydrological cycles and ecosystem food webs. Lichens are also of special interest because of their reliance on atmospheric deposition for nutrients and water, which makes them particularly sensitive to air pollution. Therefore, they are often used as bioindicators of air pollution, climate change, and general ecosystem health. This thesis examines the functional trait patterns of lichens in two contrasting regions with fundamentally different kinds of data. To better understand the patterns of lichen functional traits, we examined reproductive, morphological, and chemical trait variation along precipitation and temperature gradients in Oregon. -

H. Thorsten Lumbsch VP, Science & Education the Field Museum 1400
H. Thorsten Lumbsch VP, Science & Education The Field Museum 1400 S. Lake Shore Drive Chicago, Illinois 60605 USA Tel: 1-312-665-7881 E-mail: [email protected] Research interests Evolution and Systematics of Fungi Biogeography and Diversification Rates of Fungi Species delimitation Diversity of lichen-forming fungi Professional Experience Since 2017 Vice President, Science & Education, The Field Museum, Chicago. USA 2014-2017 Director, Integrative Research Center, Science & Education, The Field Museum, Chicago, USA. Since 2014 Curator, Integrative Research Center, Science & Education, The Field Museum, Chicago, USA. 2013-2014 Associate Director, Integrative Research Center, Science & Education, The Field Museum, Chicago, USA. 2009-2013 Chair, Dept. of Botany, The Field Museum, Chicago, USA. Since 2011 MacArthur Associate Curator, Dept. of Botany, The Field Museum, Chicago, USA. 2006-2014 Associate Curator, Dept. of Botany, The Field Museum, Chicago, USA. 2005-2009 Head of Cryptogams, Dept. of Botany, The Field Museum, Chicago, USA. Since 2004 Member, Committee on Evolutionary Biology, University of Chicago. Courses: BIOS 430 Evolution (UIC), BIOS 23410 Complex Interactions: Coevolution, Parasites, Mutualists, and Cheaters (U of C) Reading group: Phylogenetic methods. 2003-2006 Assistant Curator, Dept. of Botany, The Field Museum, Chicago, USA. 1998-2003 Privatdozent (Assistant Professor), Botanical Institute, University – GHS - Essen. Lectures: General Botany, Evolution of lower plants, Photosynthesis, Courses: Cryptogams, Biology -
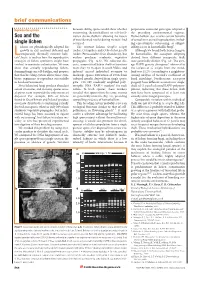
Brief Comms Layout 6/4Mx
brief communications Reproductive systems between sibling spores would show whether perpetuates successful genotypes adapted to outcrossing (heterothallism) or self-fertil- the prevailing environmental regimes. Sex and the ization (homothallism: allowing the fusion Homothallism also retains certain benefits of two identical nuclei during meiosis) had of sexual over asexual reproduction, includ- single lichen occurred. ing opportunistic outcrossing, as obligate ichens are physiologically adapted for The crustose lichens Graphis scripta selfing is rare in homothallic fungi8. growth in dry, nutrient-deficient and (order: Ostropales) and Ochrolechia parella Although we found both lichen fungi to Ltemporarily thermally extreme habi- (order: Pertusariales) fruit abundantly, but be homothallic, the ascospore offspring tats1, but it is unclear how the reproductive neither produces symbiotic vegetative derived from different conspecific thalli strategies of lichen symbionts might have propagules (Fig. 1a,b). We collected dis- were genetically distinct (Fig. 1d). The aver- evolved to maximize colonization. We now crete, symmetrical lichen thalli at locations age RAPD genetic divergence9 observed in show that sexually reproducing lichen- more than 10 m apart in south Wales, and five isolates of G. scripta from one wood- forming fungi can self-fertilize, and propose induced excised individual ascomata to land was 15.2% (according to a neighbour- that this breeding system allows these sym- discharge spores. Extraction of DNA from joining analysis of Jaccard’s coefficient of biotic organisms to reproduce successfully cultured mycelia derived from single spores band matching). Furthermore, ascospore in harsh environments. gave 218–263 randomly amplified poly- progeny from different ascomata on ‘single’ Most lichenized fungi produce abundant morphic DNA (RAPD) markers4 for each thalli of O. -

Taxonomy of Bryoria Section Implexae (Parmeliaceae, Lecanoromycetes) in North America and Europe, Based on Chemical, Morphological and Molecular Data
Ann. Bot. Fennici 51: 345–371 ISSN 0003-3847 (print) ISSN 1797-2442 (online) Helsinki 22 September 2014 © Finnish Zoological and Botanical Publishing Board 2014 Taxonomy of Bryoria section Implexae (Parmeliaceae, Lecanoromycetes) in North America and Europe, based on chemical, morphological and molecular data Saara Velmala1,*, Leena Myllys1, Trevor Goward2, Håkon Holien3 & Pekka Halonen4 1) Botanical Museum, Finnish Museum of Natural History, P.O. Box 7, FI-00014 University of Helsinki, Finland (*corresponding author’s e-mail: [email protected]) 2) UBC Herbarium, Beaty Museum, University of British Columbia, Vancouver, BC V6T 1Z4, Canada (mailing address: Enlichened Consulting Ltd., 5369 Clearwater Valley Road, Upper Clearwater, BC V0E 1N1, Canada) 3) Nord-Trøndelag University College, Serviceboks 2501, N-7729 Steinkjer, Norway 4) Botanical Museum, Department of Biology, P.O. Box 3000, FI-90014 University of Oulu, Finland Received 31 Jan. 2014, final version received 13 June 2014, accepted 18 June 2014 Velmala, S., Myllys, L., Goward, T., Holien, H. & Halonen, P. 2014: Taxonomy of Bryoria section Implexae (Parmeliaceae, Lecanoromycetes) in North America and Europe, based on chemical, morphological and molecular data. — Ann. Bot. Fennici 51: 345–371. Ninety-seven ingroup specimens of Bryoria section Implexae (Parmeliaceae, Leca- noromycetes) were studied using molecular, chemical, morphological and geographic characters. The molecular data included nuclear ribosomal markers (ITS, IGS) and the partial glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. In addition to par- simony analyses, a haplotype network was constructed. Phylogenetic analyses strongly supported the monophyly of the section Implexae. The specimens were grouped into two monophyletic clades. Clade 1 encompassed all esorediate material from North America, whereas Clade 2 included both sorediate North American material and all European material.