Selective Inhibition
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

And Prednisolone on Acute Inflammatory Response in the Rat
Ann Rheum Dis: first published as 10.1136/ard.36.2.160 on 1 April 1977. Downloaded from Annals of the Rheumatic Diseases, 1977, 36, 160-165 Comparison of the effects of alclofenac, flurbiprofen, and prednisolone on acute inflammatory response in the rat J. WOODLAND, B. VERNON-ROBERTS, K. V. SWETTENHAM, AND H. L. F. CURREY From the Bone andJoint Research Unit, The London Hospital, London El JBB SUMMARY The fluid and cellular phases of the inflammatory response were measured using a technique employing subcutaneous implantation of polyurethane foam cubes impregnated with heat-killed Mycobacterium tuberculosis. Flurbiprofen and prednisolone were equipotent and were capable of almost completely suppressing fluid and cellular responses, while alclofenac was less potent at nontoxic dose levels. Study of the patterns of cellular exudation by image analysing computer showed that alclofenac appears unique in that it produces a well-defined cell-free zone between the edge ofthe implanted cubes and a band ofneutrophil polymorphs within the cubes. A model for studying the anti-inflammatory action profen, at present being developed as a new copyright. anti- of drugs in the rat has been developed in this labora- inflammatory and analgesic agent. The effects of tory (Clarke et al., 1975) using the subcutaneous both drugs have been compared with prednisolone, implantation of polyurethane cubes impregnated a known potent anti-inflammatory drug. with an irritant. When removed 5 days after im- plantation, the cubes can be analysed separately for Materials and methods the protein content of the fluid exudate and for cellular exudation. Using this method to test a Sprague-Dawley rats weighing 250-300 g were usedhttp://ard.bmj.com/ variety of drugs used in the treatment of rheumatoid throughout. -

Analgesic Anti-Inflammatory Adhesive Preparations
Europaisches Patentamt (19) European Patent Office Office europeeneen des brevets EP 0 71 3 697 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) |nt Cl.e: A61K9/70 29.05.1996 Bulletin 1996/22 (21) Application number: 95307605.6 (22) Date of filing: 25.10.1995 (84) Designated Contracting States: • Orihara, Masamichi DE ES FR GB IT Minamisatama-gun Saitama-ken (JP) • Sugimoto, Yoshio (30) Priority: 26.10.1994 J P 284573/94 Saitama-ken (JP) • Yamazaki, Masaru (71) Applicant: Tokuhon Corporation Kitakatsushika-gun Saitama-ken (JP) Tokyo (JP) • Hoshino, Mitsunari Saitama-ken (JP) (72) Inventors: • Uchikawa, Masumasa • Sasaki, Yasuhiko Saitama-ken (JP) Saitama-ken (JP) • Arai, Hiroshi • Matsumura, Yukihiro Saitama-ken (JP) Saitama-ken (JP) • Imai, Susumu (74) Representative: Thomas, Roger Tamlyn et al Kitatsushika-gun Saitama-ken (JP) D. Young & Co. • Tooyama, Tetsuhiro 21 New Fetter Lane Saitama-ken (JP) London EC4A 1 DA (GB) (54) Analgesic anti-inflammatory adhesive preparations (57) A non-steroidal analgesic anti-inflammatory phenylyl)propionic acid and a modified copolymer in adhesive preparation obtained by coating one surface which methylmethacrylate is allowed to graft-polymer- of support with an adhesive containing S-(+)-2- ize on said copolymer, or with an adhesive containing ( 2-fluoro-4-biphenylyl)propionic acid and a styrene-iso- S-(+)-2-(2-fluoro-4-biphenylyl) propionic acid, sodium prene-styrene block copolymer or S-(+)-2-(2-fluor-4-bi- carboxymethyl cellulose and sodium polyacrylate. Is- O) CO CO o a. LU Printed by Jouve, 75001 PARIS (FR) EP 0 713 697 A1 Description The present invention concerns a non-steroidal analgesic anti-inflammatory adhesive preparation which contains as the pharmacologically effective component S-(+)-2-(2-fluoro-4-biphenylyl) propionic acid [abbreviated S-(+)-FP 5 hereinafter] which may be optically resolved from the racemic form 2-(2-fluoro-4-biphenylyl) propionic acid (also called: flurbiprofen). -

United States Patent (19) 11 Patent Number: 5,192,753 Mcgeer Et Al
USOO5192753A United States Patent (19) 11 Patent Number: 5,192,753 McGeer et al. 45 Date of Patent: Mar. 9, 1993 54 ANTI-RHEUMATOID ARTHRITIC DRUGS mentia: Prevalance and Incidence', In: B. Reisberg (ed) N THE TREATMENT OF DEMENTIA Alzheimer's Disease, The Free Press, 1983, 141-148. 76 Inventors: Patrick L. McGeer, 4727 West 2nd R. Sulkava, J. Wikstrom, A. Aromaa et al., "Prevalence Ave., Vancouver, B. C., Canada, of Severe Dementia in Finland’, Neurology 1985; 35: V6T Cl; Joseph Rogers, 7646 W. 1025-1029. Julie Dr., Glendale, Ariz. 85308; S. Itagaki, P. L. McGeer, H. Akiyama, "Presence of John Sibley, 87 Leddy Crescent, T-cytotoxic Suppressor and Leucocyte Common Anti Saskatoon, Saskatchewan, Canada, gen Positive Cells in Alzheimer's Disease Brain Tissue", S7H 3Y9; Edith McGeer, 4727 West Neuroscience Letters 1988; 91: 259-264. 2nd Ave., Vancouver, B.C., Canada, J. Rogers, J. Luber-Narod, S. D. Styren, and W. H. V6T 1C Civin, "Expression of Immune System-Associated An tigens By Cells of the Human Central Nervous System: (21) Appl. No.: 689,499 Relationship to the Pathology of Alzheimer's Disease" (22 Filed: Apr. 23, 1991 Neurobiology of Aging, 1988, vol. 9. 339-349. 51 Int. Cl. ................... A61K 31/60; A61K 31/615; P. L. McGeer, H. Akiyama, S. Itagaki, and E. G. A61K 31/54; A61K 31/44; A61K 3/425; McGeer, "Immune System Response in Alzheimer's A61K 31/42; A61K 31/415, A61K 31/40; Disease', The Canadian Journal of Neurological Sci A61K 31/38; A61K 31/195; A61K 31/19 ences 1989; 16:56-527. -

Pharmaceutical Compositions Containing Non-Steroidal Anti-Inflammatory Drugs
Europaisches Patentamt J European Patent Office © Publication number: 0 259 047 Office europeen des brevets A1 s EUROPEAN PATENT APPLICATION © Application number: 87307246.6 © int. CIA A61K 45/06 A61 , K 31/60 , A61K 31/405, © Date of filing: 17.0&87 //(A61 K31/60,31 :1 85),(A61 K31/- 405,31:185) ® Priority: 18.08.86 GB 8620073 © Applicant: DELANDALE LABORATORIES LIMITED © Date of publication of application: Delandale House 37 Old Dover Road 09.03.88 Bulletin 88/10 Canterbury Kent CT1 3JF(GB) © Designated Contracting States: @ inventor: Trigger, David John AT BE CH DE ES FR GB GR IT LI LU NL SE 74 Mountbatten Way Brabourne Lees Ashford Kent TN25 6PU(GB) © Representative: Paget, Hugh Charles Edward etal MEWBURN ELUS & CO. 2/3 Cursitor Street London EC4A 1BQ(GB) © Pharmaceutical compositions containing non-steroidal anti-Inflammatory drugs. © A pharmaceutical composition for treating an inflammatory condition contains a non-steroidal anti-inflam- matory drug (NSAID) and a physiologically acceptable aromatic hydroxysulphonic acid. The two components may be for simultaneousr separate or sequential use. Administration of the two components, for example of aspirin with ethamsylate, reduces the damage which the NSAID does to the gastrointestinal tract while having no deleterious effect on the anti-inflammatory activity. s in cm Q. LU Xerox Copy Centre 0 259 047 PHARMACEUTICAL COMPOSITIONS CONTAINING NON-STEROIDAL ANTI-INFLAMMATORY DRUGS The present invention relates to pharmaceutical compositions containing aspirin or other non-steroidal anti-inflammatory drugs (NSAIDs). The non-steroidal anti-inflammatory drugs are a group of compounds which inhibit the biosynthesis of prostaglandins. -

Oral Nonsteroidal Anti-Inflammatory Drugs for Neuropathic Pain
Oral nonsteroidal anti-inflammatory drugs for neuropathic pain (Review) Moore RA, Chi CC, Wiffen PJ, Derry S, Rice ASC This is a reprint of a Cochrane review, prepared and maintained by The Cochrane Collaboration and published in The Cochrane Library 2015, Issue 10 http://www.thecochranelibrary.com Oral nonsteroidal anti-inflammatory drugs for neuropathic pain (Review) Copyright © 2015 The Cochrane Collaboration. Published by John Wiley & Sons, Ltd. TABLE OF CONTENTS HEADER....................................... 1 ABSTRACT ...................................... 1 PLAINLANGUAGESUMMARY . 2 BACKGROUND .................................... 2 OBJECTIVES ..................................... 4 METHODS ...................................... 4 RESULTS....................................... 7 Figure1. ..................................... 8 Figure2. ..................................... 9 DISCUSSION ..................................... 10 AUTHORS’CONCLUSIONS . 11 ACKNOWLEDGEMENTS . 11 REFERENCES ..................................... 12 CHARACTERISTICSOFSTUDIES . 16 DATAANDANALYSES. 20 APPENDICES ..................................... 20 WHAT’SNEW..................................... 25 HISTORY....................................... 25 CONTRIBUTIONSOFAUTHORS . 25 DECLARATIONSOFINTEREST . 26 SOURCESOFSUPPORT . 26 DIFFERENCES BETWEEN PROTOCOL AND REVIEW . .... 26 NOTES........................................ 26 Oral nonsteroidal anti-inflammatory drugs for neuropathic pain (Review) i Copyright © 2015 The Cochrane Collaboration. Published by John Wiley -

(CD-P-PH/PHO) Report Classification/Justifica
COMMITTEE OF EXPERTS ON THE CLASSIFICATION OF MEDICINES AS REGARDS THEIR SUPPLY (CD-P-PH/PHO) Report classification/justification of - Medicines belonging to the ATC group M01 (Antiinflammatory and antirheumatic products) Table of Contents Page INTRODUCTION 6 DISCLAIMER 8 GLOSSARY OF TERMS USED IN THIS DOCUMENT 9 ACTIVE SUBSTANCES Phenylbutazone (ATC: M01AA01) 11 Mofebutazone (ATC: M01AA02) 17 Oxyphenbutazone (ATC: M01AA03) 18 Clofezone (ATC: M01AA05) 19 Kebuzone (ATC: M01AA06) 20 Indometacin (ATC: M01AB01) 21 Sulindac (ATC: M01AB02) 25 Tolmetin (ATC: M01AB03) 30 Zomepirac (ATC: M01AB04) 33 Diclofenac (ATC: M01AB05) 34 Alclofenac (ATC: M01AB06) 39 Bumadizone (ATC: M01AB07) 40 Etodolac (ATC: M01AB08) 41 Lonazolac (ATC: M01AB09) 45 Fentiazac (ATC: M01AB10) 46 Acemetacin (ATC: M01AB11) 48 Difenpiramide (ATC: M01AB12) 53 Oxametacin (ATC: M01AB13) 54 Proglumetacin (ATC: M01AB14) 55 Ketorolac (ATC: M01AB15) 57 Aceclofenac (ATC: M01AB16) 63 Bufexamac (ATC: M01AB17) 67 2 Indometacin, Combinations (ATC: M01AB51) 68 Diclofenac, Combinations (ATC: M01AB55) 69 Piroxicam (ATC: M01AC01) 73 Tenoxicam (ATC: M01AC02) 77 Droxicam (ATC: M01AC04) 82 Lornoxicam (ATC: M01AC05) 83 Meloxicam (ATC: M01AC06) 87 Meloxicam, Combinations (ATC: M01AC56) 91 Ibuprofen (ATC: M01AE01) 92 Naproxen (ATC: M01AE02) 98 Ketoprofen (ATC: M01AE03) 104 Fenoprofen (ATC: M01AE04) 109 Fenbufen (ATC: M01AE05) 112 Benoxaprofen (ATC: M01AE06) 113 Suprofen (ATC: M01AE07) 114 Pirprofen (ATC: M01AE08) 115 Flurbiprofen (ATC: M01AE09) 116 Indoprofen (ATC: M01AE10) 120 Tiaprofenic Acid (ATC: -

Pharmaceuticals (Monocomponent Products) ………………………..………… 31 Pharmaceuticals (Combination and Group Products) ………………….……
DESA The Department of Economic and Social Affairs of the United Nations Secretariat is a vital interface between global and policies in the economic, social and environmental spheres and national action. The Department works in three main interlinked areas: (i) it compiles, generates and analyses a wide range of economic, social and environmental data and information on which States Members of the United Nations draw to review common problems and to take stock of policy options; (ii) it facilitates the negotiations of Member States in many intergovernmental bodies on joint courses of action to address ongoing or emerging global challenges; and (iii) it advises interested Governments on the ways and means of translating policy frameworks developed in United Nations conferences and summits into programmes at the country level and, through technical assistance, helps build national capacities. Note Symbols of United Nations documents are composed of the capital letters combined with figures. Mention of such a symbol indicates a reference to a United Nations document. Applications for the right to reproduce this work or parts thereof are welcomed and should be sent to the Secretary, United Nations Publications Board, United Nations Headquarters, New York, NY 10017, United States of America. Governments and governmental institutions may reproduce this work or parts thereof without permission, but are requested to inform the United Nations of such reproduction. UNITED NATIONS PUBLICATION Copyright @ United Nations, 2005 All rights reserved TABLE OF CONTENTS Introduction …………………………………………………………..……..……..….. 4 Alphabetical Listing of products ……..………………………………..….….…..….... 8 Classified Listing of products ………………………………………………………… 20 List of codes for countries, territories and areas ………………………...…….……… 30 PART I. REGULATORY INFORMATION Pharmaceuticals (monocomponent products) ………………………..………… 31 Pharmaceuticals (combination and group products) ………………….……........ -
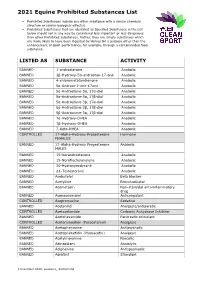
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

Anti-Inflammatory/Analgesic Combination of Alpha
Patentamt JEuropàischesEuropean Patent Office ® Publication number: 0 1 09 036 Office européen des brevets g "j © EUROPEAN PATENT SPECIFICATION (45) Dateof publication of patent spécification: 02.09.87 (jjj) jnt ci 4- A 61 K 31/415, A 61 K 31/62, (22) Dateoffiling: 08.11.83 (54) Anti-inf lammatory/analgesic combination of alpha-fluoromethylhistidine and a selected non-steroidal anti-inflammatory drug (NSAID). (§) Priority: 15.11.82 US 441581 (73) Proprietor: MERCK & CO. INC. 126, East Lincoln Avenue P.O. Box 2000 Rahway New Jersey 07065 (US) (43) Date of publication of application: 23.05.84 Bulletin 84/21 @ Inventor: Goldenberg, Marvin M. 721 Shackamaxon Drive (45) Publication of the grant of the patent: Westfield New Jersey 07090 (US) 02.09.87 Bulletin 87/36 (74) Representative: Abitz, Walter, Dr.-lng. et al (84) Designated Contracting States: Abitz, Morf, Gritschneder, Freiherr von CH DE FR GB IT LI NL Wittgenstein Postfach 86 01 09 D-8000 Munchen 86 (DE) (§) References cited: EP-A-0 046290 US-A-4325 961 CÛ (£> o o> o Note: Within nine months from the publication of the mention of the grant of the European patent, any person may give notice to the European Patent Office of opposition to the European patent granted. Notice of opposition shall CL be filed in a written reasoned statement. It shall not be deemed to have been filed until the opposition fee has been LU paid. (Art. 99( 1 ) European patent convention). Courier Press, Leamington Spa, England. BACKGROUND OF THE INVENTION This invention relates to novel pharmaceutical combinations comprising a-fluoromethylhistidine (FMH) and a non-steroidal anti-inflammatory/analgesic drug (NSAID) particularly indomethacin, diflunisal and naproxen. -

Plants As Sources of Anti-Inflammatory Agents
molecules Review Plants as Sources of Anti-Inflammatory Agents Clara dos Reis Nunes 1 , Mariana Barreto Arantes 1, Silvia Menezes de Faria Pereira 1, Larissa Leandro da Cruz 1, Michel de Souza Passos 2, Luana Pereira de Moraes 1, Ivo José Curcino Vieira 2 and Daniela Barros de Oliveira 1,* 1 Laboratório de Tecnologia de Alimentos, Centro de Ciências e Tecnologias Agropecuárias, Universidade Estadual do Norte Fluminense Darcy Ribeiro, Campos dos Goytacazes, RJ 28013-602, Brazil; [email protected] (C.d.R.N.); [email protected] (M.B.A.); [email protected] (S.M.d.F.P.); [email protected] (L.L.d.C.); [email protected] (L.P.d.M.) 2 Laboratório de Ciências Químicas, Centro de Ciências e Tecnologia, UniversidadeEstadual do Norte Fluminense Darcy Ribeiro, Campos dos Goytacazes, RJ 28013-602, Brazil; [email protected] (M.d.S.P.); [email protected] (I.J.C.V.) * Correspondence: [email protected]; Tel.: +55-22-988395160 Academic Editors: Thea Magrone, Rodrigo Valenzuela and Karel Šmejkal Received: 29 June 2020; Accepted: 5 August 2020; Published: 15 August 2020 Abstract: Plants represent the main source of molecules for the development of new drugs, which intensifies the interest of transnational industries in searching for substances obtained from plant sources, especially since the vast majority of species have not yet been studied chemically or biologically, particularly concerning anti-inflammatory action. Anti-inflammatory drugs can interfere in the pathophysiological process of inflammation, to minimize tissue damage and provide greater comfort to the patient. Therefore, it is important to note that due to the existence of a large number of species available for research, the successful development of new naturally occurring anti-inflammatory drugs depends mainly on a multidisciplinary effort to find new molecules. -
Poison Or Antibiotic? a Guide to "Class" Entries
Poison or Antibiotic? A Guide to “Class” Entries Preface Most entries in the Poisons List, i.e. the Schedule 10, and the Schedules 1, 2 and 3 to the Pharmacy and Poisons Regulations (Cap. 138A) are in the form of individual drugs and their salts, e.g. “Abacavir; its salts”. However, some entries are in the form of a “class”, e.g. “Barbituric acid; its salts; its derivatives …”. In such cases, it is not always clear which drugs are members of the class (e.g. amobarbital, barbital, pentobarbital, phenobarbital, etc. are poisons, being derivatives of barbituric acid). Likewise, the Antibiotics Ordinance (Cap. 137) applies to the substances specified in Schedule 1 to the Antibiotics Regulations, to their salts and derivatives, and to the salts of such derivatives. Again, it is not always clear which drugs are derivatives of an antibiotic named in the Schedule (e.g. demeclocycline, doxycycline, tigecycline, etc. are antibiotics, being derivatives of “Tetracycline” named in the Schedule). This Guide provides a list of such drugs which are available as registered pharmaceutical products in Hong Kong. Drugs which are not available as registered pharmaceutical products in Hong Kong are also included in this Guide as far as possible. It should be noted that it is not possible to compile a complete list of all these drugs, simply because there is no limit to the number of “derivatives” a parent chemical can have. This Guide should be read in conjunction with the Schedules 1, 2, 3, and 10 to the Pharmacy and Poisons Regulations, and Schedule 1 to the Antibiotics Regulations, if the poison/antibiotic classification of a particular pharmaceutical product is to be determined. -

High Daily Dose and Being a Substrate of Cytochrome P450 Enzymes Are Two Important Predictors of Drug-Induced Liver Injury
DMD Fast Forward. Published on January 24, 2014 as DOI: 10.1124/dmd.113.056267 DMD FastThis Forward.article has not Published been copyedited on and January formatted. 24, The 2014final version as doi:10.1124/dmd.113.056267 may differ from this version. DMD #56267 High daily dose and being a substrate of Cytochrome P450 enzymes are two important predictors of drug-induced liver injury Ke Yu, Xingchao Geng, Minjun Chen, Jie Zhang, Bingshun Wang, Katarina Ilic and Weida Tong Division of Bioinformatics and Biostatistics, National Center for Toxicological Research, Downloaded from US Food and Drug Administration, Jefferson, AR, USA (K.Y., M.C., J.Z., W.T) National Center for Safety Evaluation of Drugs, National Institute for Food and Drug Control, China's State Food and Drug Administration, Beijing, China (X.G.) dmd.aspetjournals.org Department of Biostatistics, Shanghai Jiao Tong University School of Medicine, Shanghai, China (B.W.) Department of Pharmacology, Faculty of Pharmacy, University of Belgrade, Belgrade, Serbia (K.I.) at ASPET Journals on September 26, 2021 1 Copyright 2014 by the American Society for Pharmacology and Experimental Therapeutics. DMD Fast Forward. Published on January 24, 2014 as DOI: 10.1124/dmd.113.056267 This article has not been copyedited and formatted. The final version may differ from this version. DMD #56267 Running title: Two important predictors for drug-induced liver injury Corresponding authors: Katarina Ilic, Department of Pharmacology, Faculty of Pharmacy, University of Belgrade, 450 Vojvode Stepe, Belgrade 11221, Serbia. E-mail address: [email protected]. Weida Tong, Division of Bioinformatics and Biostatistics, National Center for Toxicological Research, US Food and Drug Administration, 3900 NCTR Road, Jefferson, Downloaded from AR 72079, USA.