And Studies of the Exotic Cotton Leafroll Dwarf Virus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Normas Para Confecção Da Versão
UNIVERSIDADE FEDERAL DE UBERLÂNDIA INSTITUTO DE GENÉTICA E BIOQUÍMICA PÓS-GRADUAÇÃO EM GENÉTICA E BIOQUÍMICA Poliploidia e variações reprodutivas em Bombacoideae (Malvaceae): distribuição geográfica, filogeografia e tamanho do genoma Aluna: Rafaela Cabral Marinho Orientadora: Profª. Drª. Ana Maria Bonetti Co-orientador: Prof. Dr. Paulo Eugênio Alves Macedo de Oliveira UBERLÂNDIA - MG 2017 UNIVERSIDADE FEDERAL DE UBERLÂNDIA INSTITUTO DE GENÉTICA E BIOQUÍMICA PÓS-GRADUAÇÃO EM GENÉTICA E BIOQUÍMICA Poliploidia e variações reprodutivas em Bombacoideae (Malvaceae): distribuição geográfica, filogeografia e tamanho do genoma Aluna: Rafaela Cabral Marinho Orientadora: Profª. Drª. Ana Maria Bonetti Co-orientador: Prof. Dr. Paulo Eugênio Alves Macedo de Oliveira Tese apresentada à Universidade Federal de Uberlândia como parte dos requisitos para obtenção do Título de Doutora em Genética e Bioquímica (Área Genética) UBERLÂNDIA – MG 2017 ii Dados Internacionais de Catalogação na Publicação (CIP) Sistema de Bibliotecas da UFU, MG, Brasil. M338p Marinho, Rafaela Cabral, 1988 2017 Poliploidia e variações reprodutivas em Bombacoideae (Malvaceae): distribuição geográfica, filogeografia e tamanho do genoma / Rafaela Cabral Marinho. - 2017. 100 f. : il. Orientadora: Ana Maria Bonetti. Coorientador: Paulo Eugênio Alves Macedo de Oliveira. Tese (doutorado) - Universidade Federal de Uberlândia, Programa de Pós-Graduação em Genética e Bioquímica. Disponível em: http://dx.doi.org/10.14393/ufu.di.2018.134 Inclui bibliografia. 1. Genética - Teses. 2. Malvaceae -

Yarra Yarra Group Inc (Incorporation No
Australian Plants Society Yarra Yarra Group Inc (Incorporation No. A0039676Y) Newsletter May 2018 May 3: Maree & Graham Goods : Gardening in the Wimmera Graham & Maree come from a farming and administration background. They are Life Members of the Wimmera Growers of Australian Plants of which they have been members for over 40 years. Their garden has been open to the public on several occasions for various charities. They ran a wholesale nursery for several years specialising in Eremophilas. Graham and Maree’s current project is growing plants, landscaping and overseeing the planting of the gardens surrounding the new building of their Church which was opened recently. They first became involved with Australia’s inland deserts in 2002 with an organisation called Desert Discovery as part of the Botany team. In 2008, Maree became the Botany Team Leader, while Graham was involved with the photography and identification of specimens. They have done volunteer work for Australian Wildlife Conservancy, Friends of the Great Victoria Desert, Victorian and Western Australian Herbariums in collecting, identifying and processing specimens. Graham and Maree are co-authors of the book, Birds and Plants of the Little Desert, a photographic guide and Maree a co-author of Australia’s Eremophilas Website: apsyarrayarra.org.au Facebook: facebook.com/APSYarraYarra Email: [email protected] | 1 APS Yarra Yarra Particulars APS YY General Meeting APS YY Garden Visits: Speakers: May 13, 2 pm, Bill Aitchison and Sue Guymer Garden, Donvale. 7-June Greg Moore Urban Greening 5-July Ryan Phillips Animal Interactions Natural bush garden with ponds,stream and large dam merging to adjacent bush. -

Comparative Transcriptomic Analysis to Identify the Genes Related to Delayed Gland Morphogenesis in Gossypium Bickii
G C A T T A C G G C A T genes Article Comparative Transcriptomic Analysis to Identify the Genes Related to Delayed Gland Morphogenesis in Gossypium bickii Mushtaque Ali 1, Hailiang Cheng 1, Mahtab Soomro 1, Li Shuyan 1, Muhammad Bilal Tufail 1, Mian Faisal Nazir 1 , Xiaoxu Feng 1,2, Youping Zhang 1, Zuo Dongyun 1, Lv Limin 1, Qiaolian Wang 1 and Guoli Song 1,* 1 State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang 455000, China; [email protected] (M.A.); [email protected] (H.C.); [email protected] (M.S.); [email protected] (L.S.); [email protected] (M.B.T.); [email protected] (M.F.N.); [email protected] (X.F.); [email protected] (Y.Z.); [email protected] (Z.D.); [email protected] (L.L.); [email protected] (Q.W.) 2 Plant Genetics, Gambloux Agro Bio Tech, University of Liege, 5030 Gambloux, Belgium * Correspondence: [email protected]; Tel.: +86-3722562377 Received: 20 March 2020; Accepted: 19 April 2020; Published: 26 April 2020 Abstract: Cotton is one of the major industrial crops that supply natural fibers and oil for industries. This study was conducted to understand the mechanism of delayed gland morphogenesis in seeds of Gossypium bickii. In this study, we compared glandless seeds of G. bickii with glanded seeds of Gossypium arboreum. High-throughput sequencing technology was used to explore and classify the expression patterns of gland-related genes in seeds and seedlings of cotton plants. Approximately 131.33 Gigabases of raw data from 12 RNA sequencing samples with three biological replicates were generated. -

Growing Australian Plants
ASSOqIATTON OF SOCIETIES FOR GROWING AUSTRALIAN PLANTS LETTER NO 11 :ISSN: 1488-1488 at Hibiscus species H. forsterii F.D. Wilson (Sect. Furcaria DC') Contaitrer srown guclerim (jueensland 6n May 2007 Images C' Harve) G. Harvey Hibiscus diversifolius winter bloom 2nd June 2007. lnage NL 1l p.l Welcome to Newsletter No I l The front page depicts H. forsterii from the Cape York region of Queensland. The image of H. diversifolius blooms at the foot of the page contrast sharply with the purple flowers obtained in summer ; (see front page of Newsletter No 10). When the wirrter temperature is not quite so cold the off-white colour seen in the image, tends towards pale lemon and with still warmer weather becomes pink and finally red/purple in summer. It is being sold in Fairhill Native Plant Nursery as "Colour Magic". The so called'Norfolk Island Hibiscus" : Lagunaria patersonius (Andrews) G. Don subsp. patersonius, has appeared on a Norfolk Island postage stamp as illustrated bottom right on the front page. Winter arrived with a vengeance in the first week of June following a very mild autumn. Many Hibiscus came into bud early and H. heterophyllus are now in full bloom on the nofthern end of the Sunshine Coast. Dion Harrison reports some early blooms from the Mount Crosby Cliffs and Kenmore near Brisbane. At Buderim our rainfall for June was 211mm, our average for that month being 14lmm. Unfortunately this coastal rain hasn't penetrated inland to much extent, where it is much needed in the dam catchment areas. H. -

Antisense Suppression of a (+)-D-Cadinene Synthase Gene In
Antisense Suppression of a (1)-d-Cadinene Synthase Gene in Cotton Prevents the Induction of This Defense Response Gene during Bacterial Blight Infection But Not Its Constitutive Expression1[w] Belinda J. Townsend2, Andrew Poole, Christopher J. Blake, and Danny J. Llewellyn* Commonwealth Scientific and Industrial Research Organisation-Plant Industry, Canberra, Australian Capital Territory 2601, Australia (B.J.T., A.P., D.J.L.); and Department of Biochemistry and Molecular Biology (B.J.T.) and Research School of Chemistry (C.J.B.), Australian National University, Canberra, Australian Capital Territory 0200, Australia In cotton (Gossypium hirsutum) the enzyme (1)-d-cadinene synthase (CDNS) catalyzes the first committed step in the biosynthesis of cadinane-type sesquiterpenes, such as gossypol, that provide constitutive and inducible protection against pests and diseases. A cotton cDNA clone encoding CDNS (cdn1-C4) was isolated from developing embryos and functionally characterized. Southern analysis showed that CDNS genes belong to a large multigene family, of which five genomic clones were studied, including three pseudogenes and one gene that may represent another subfamily of CDNS. CDNS expression was shown to be induced in cotton infected with either the bacterial blight or verticillium wilt pathogens. Constructs for the constitutive or seed-specific antisense suppression of cdn1-C4 were introduced into cotton by Agrobacterium-mediated transformation. Gossypol levels were not reduced in the seeds of transformants with either construct, nor was the induction of CDNS expression affected in stems of the constitutive antisense plants infected with Verticillium dahliae Kleb. However, the induction of CDNS mRNA and protein in response to bacterial blight infection of cotyledons was completely blocked in the constitutive antisense plants. -
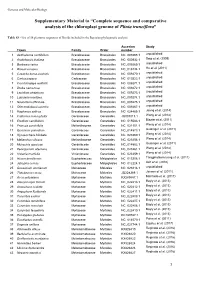
Complete Sequence and Comparative Analysis of the Chloroplast Genome of Plinia Trunciflora”
Genetics and Molecular Biology Supplementary Material to “Complete sequence and comparative analysis of the chloroplast genome of Plinia trunciflora” Table S3 - List of 56 plastome sequences of Rosids included in the Bayesian phylogenetic analysis. Accesion Study Taxon Family Order number 1 Aethionema cordifolium Brassicaceae Brassicales NC_009265.1 unpublished 2 Arabidopsis thaliana Brassicaceae Brassicales NC_000932.1 Sato et al. (1999) 3 Barbarea verna Brassicaceae Brassicales NC_009269.1 unpublished 4 Brassica napus Brassicaceae Brassicales NC_016734.1 Hu et al. (2011) 5 Capsella bursa-pastoris Brassicaceae Brassicales NC_009270.1 unpublished 6 Carica papaya Caricaceae Brassicales NC_010323.1 unpublished 7 Crucihimalaya wallichii Brassicaceae Brassicales NC_009271.1 unpublished 8 Draba nemorosa Brassicaceae Brassicales NC_009272.1 unpublished 9 Lepidium virginicum Brassicaceae Brassicales NC_009273.1 unpublished 10 Lobularia maritima Brassicaceae Brassicales NC_009274.1 unpublished 11 Nasturtium officinale Brassicaceae Brassicales NC_009275.1 unpublished 12 Olimarabidopsis pumila Brassicaceae Brassicales NC_009267.1 unpublished 13 Raphanus sativus Brassicaceae Brassicales NC_024469.1 Jeong et al. (2014) 14 California macrophylla Geraniaceae Geraniales JQ031013.1 Weng et al. (2014) 15 Erodium carvifolium Geraniaceae Geraniales NC_015083.1 Blazier et al. (2011) 16 Francoa sonchifolia Melianthaceae Geraniales NC_021101.1 Weng et al. (2014) 17 Geranium palmatum Geraniaceae Geraniales NC_014573.1 Guisinger et al. (2011) 18 Hypseocharis bilobate Geraniaceae Geraniales NC_023260.1 Weng et al. (2014) 19 Melianthus villosus Melianthaceae Geraniales NC_023256.1 Weng et al. (2014) 20 Monsonia speciose Geraniaceae Geraniales NC_014582.1 Guisinger et al. (2011) 21 Pelargonium alternans Geraniaceae Geraniales NC_023261.1 Weng et al. (2014) 22 Viviania marifolia Vivianiaceae Geraniales NC_023259.1 Weng et al. (2014) 23 Hevea brasiliensis Euphorbiaceae Malpighiales NC_015308.1 Tangphatsornruang et al. -

Lose the Plot: Cost-Effective Survey of the Peak Range, Central Queensland
Lose the plot: cost-effective survey of the Peak Range, central Queensland. Don W. Butlera and Rod J. Fensham Queensland Herbarium, Environmental Protection Agency, Mt Coot-tha Botanic Gardens, Mt Coot-tha Road, Toowong, QLD, 4066 AUSTRALIA. aCorresponding author, email: [email protected] Abstract: The Peak Range (22˚ 28’ S; 147˚ 53’ E) is an archipelago of rocky peaks set in grassy basalt rolling-plains, east of Clermont in central Queensland. This report describes the flora and vegetation based on surveys of 26 peaks. The survey recorded all plant species encountered on traverses of distinct habitat zones, which included the ‘matrix’ adjacent to each peak. The method involved effort comparable to a general flora survey but provided sufficient information to also describe floristic association among peaks, broad habitat types, and contrast vegetation on the peaks with the surrounding landscape matrix. The flora of the Peak Range includes at least 507 native vascular plant species, representing 84 plant families. Exotic species are relatively few, with 36 species recorded, but can be quite prominent in some situations. The most abundant exotic plants are the grass Melinis repens and the forb Bidens bipinnata. Plant distribution patterns among peaks suggest three primary groups related to position within the range and geology. The Peak Range makes a substantial contribution to the botanical diversity of its region and harbours several endemic plants among a flora clearly distinct from that of the surrounding terrain. The distinctiveness of the range’s flora is due to two habitat components: dry rainforest patches reliant upon fire protection afforded by cliffs and scree, and; rocky summits and hillsides supporting xeric shrublands. -

Evolution of Telomerase RNA by Dhenugen Logeswaran A
Evolution of Telomerase RNA by Dhenugen Logeswaran A Dissertation Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy Approved November 2019 by the Graduate Supervisory Committee: Julian Chen, Chair Giovanna Ghirlanda Chad Borges ARIZONA STATE UNIVERSITY December 2019 ABSTRACT The highly specialized telomerase ribonucleoprotein enzyme is composed minimally of telomerase reverse transcriptase (TERT) and telomerase RNA (TR) for catalytic activity. Telomerase is an RNA-dependent DNA polymerase that syntheizes DNA repeats at chromosome ends to maintain genome stability. While TERT is highly conserved among various groups of species, the TR subunit exhibits remarkable divergence in primary sequence, length, secondary structure and biogenesis, making TR identification extremely challenging even among closely related groups of organisms. A unique computational approach combined with in vitro telomerase activity reconstitution studies was used to identify 83 novel TRs from 10 animal kingdom phyla spanning 18 diverse classes from the most basal sponges to the late evolving vertebrates. This revealed that three structural domains, pseudoknot, a distal stem-loop moiety and box H/ACA, are conserved within TRs from basal groups to vertebrates, while group- specific elements emerge or disappear during animal TR evolution along different lineages. Next the corn-smut fungus Ustilago maydis TR was identified using an RNA- immunoprecipitation and next-generation sequencing approach followed by computational identification of TRs from 19 additional class Ustilaginomycetes fungi, leveraging conserved gene synteny among TR genes. Phylogenetic comparative analysis, in vitro telomerase activity and TR mutagenesis studies reveal a secondary structure of TRs from higher fungi, which is also conserved with vertebrates and filamentous fungi, providing a crucial link in TR evolution within the opisthokonta super-kingdom. -

51729 ANBG Friends Nletter
Friends of the Australian National Botanic Gardens NEWSLETTER Number 60 November 2008 Lift Out: What‛s out in the Gardens & What‛s on at the Gardens A happier prospect Alan Munns, President, Friends of ANBG. At the time the last Newsletter went to press I was pessimistic about the Gardens budget for 2008-09. At that time we knew that Parks Australia had not done well in the budget and I worried that the Gardens might face yet more staff cuts. Things turned out better than I feared. The Gardens got a small increase in its budget which enabled some new staff to be engaged to help with review of the management plan, asset management and the education program. The Friends Patron Mrs Marlena Jeffery welcome this development, and will continue to press for more resources for the President Alan Munns Gardens. Vice President Barbara Podger Secretary David Coutts Friends’ springtime activities this year have been very popular. Treasurer Beverley Fisher Membership Secretary Barbara Scott We provided funds to support the creation of a Spring Public Officer David Coutts Display of flowering annual plants—daisies and Sturt’s General Committee Don Beer Desert Pea. Guided spring flower walks followed by morn- John Connolly ing tea at Hudsons Café were very popular. We hope this Les Fielke Louise Muir will become an annual event. Warwick Wright Activities Coordinator Warwick Wright Anne Rawson Photo by Social events: Louise Muir Breakfast with the Birds has attracted large numbers of Newsletter Committee Margaret Clarke visitors to early-morning guided walks led by enthusias- Barbara Podger Anne Rawson tic birdos, followed by breakfast at Hudsons Café. -

Gossypium Herbaceum and G
RESEARCH ARTICLE A New Synthetic Allotetraploid (A1A1G2G2) between Gossypium herbaceum and G. australe: Bridging for Simultaneously Transferring Favorable Genes from These Two Diploid Species into Upland Cotton Quan Liu☯, Yu Chen☯, Yu Chen¤, Yingying Wang, Jinjin Chen, Tianzhen Zhang, a11111 Baoliang Zhou* State Key Laboratory of Crop Genetics & Germplasm Enhancement, MOE Hybrid Cotton R&D Engineering Research Center, Nanjing Agricultural University, Nanjing, Jiangsu, People’s Republic of China ☯ These authors contributed equally to this work. ¤ Current address: Key Laboratory of Cotton Breeding and Cultivation in Huang-Huai-Hai Plain, Ministry of Agriculture, Cotton Research Center of Shandong Academy of Agricultural Sciences, Jinan, Shandong, OPEN ACCESS People’s Republic of China * [email protected] Citation: Liu Q, Chen Y, Chen Y, Wang Y, Chen J, Zhang T, et al. (2015) A New Synthetic Allotetraploid (A1A1G2G2) between Gossypium herbaceum and G. australe: Bridging for Simultaneously Transferring Abstract Favorable Genes from These Two Diploid Species into Upland Cotton. PLoS ONE 10(4): e0123209. Gossypium herbaceum, a cultivated diploid cotton species (2n = 2x = 26, A1A1), has favor- doi:10.1371/journal.pone.0123209 able traits such as excellent drought tolerance and resistance to sucking insects and leaf Academic Editor: Jean-Marc Lacape, CIRAD, curl virus. G. australe, a wild diploid cotton species (2n = 2x = 26, G2G2), possesses numer- FRANCE ous economically valuable characteristics such as delayed pigment gland morphogenesis Received: November 20, 2014 (which is conducive to the production of seeds with very low levels of gossypol as a potential Accepted: March 1, 2015 food source for humans and animals) and resistance to insects, wilt diseases and abiotic stress. -

Gossypium Hirsutum Source
Gossypium hirsutum Source: AGRICOLA database (1970-1996) References (Biological Abstracts 1988-2000): Agrawal, A. A. (2000). Host-range evolution: Adaptation and trade-offs in fitness of mites on alternative hosts. Ecology Washington D C. [print] February 81(2): 500-508. {a} Department of Botany, University of Toronto, 25 Wilcocks Avenue, Toronto, ON, M5S 3B2, Canada Agrawal, A. A., R. Karban, et al. (2000). How leaf domatia and induced plant resistance affect herbivores, natural enemies and plant performance. Oikos . April 89(1): 70-80. {a} Dept of Botany, Univ. of Toronto, 25 Willcocks Street, Toronto, ON, M5S 3B2, Canada Ahuja, S. L. and S. K. Banerjee (2000). Stability for bollworm resistance, jassid grade, seed cotton yield and its components of cytotypes in cotton (Gossypium hirsutum L.). Indian Journal of Agricultural Research. [print] June 34(2): 71-77. {a} Central Institute for Cotton Research Regional Station, Sirsa, 125055, India Anadranistakis, M., A. Liakatas, et al. (2000). Crop water requirements model tested for crops grown in Greece. Agricultural Water Management. [print] August 45(3): 297-316. {a} Agricultural University of Athens, 75 Iera Odos, GR-118 55, Athens, Greece Andersland, J. M. and B. A. Triplett (2000). Selective extraction of cotton fiber cytoplasts to identify cytoskeletal-associated proteins. Plant Physiology and Biochemistry Paris. March 38(3): 193-199. {a} ARS, Southern Regional Research Center, USDA, 1100 Robert E. Lee Blvd., New Orleans, LA, 70124-4305, USA Ashraf, M. and S. Ahmad (2000). Influence of sodium chloride on ion accumulation, yield components and fibre characteristics in salt-tolerant and salt-sensitive lines of cotton (Gossypium hirsutum L.). -

Resistant Germplasm in Gossypium Species and Related Plants to the Reniform Nematode
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1981 Resistant Germplasm in Gossypium Species and Related Plants to the Reniform Nematode. Choi-pheng Yik Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Yik, Choi-pheng, "Resistant Germplasm in Gossypium Species and Related Plants to the Reniform Nematode." (1981). LSU Historical Dissertations and Theses. 3622. https://digitalcommons.lsu.edu/gradschool_disstheses/3622 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. INFORMATION TO USERS This was produced from a copy of a document sent to us for microfilming. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help you understand markings or notations which may appear on this reproduction. 1.The sign or “target” for pages apparently lacking from the document photographed is “Missing Page(s)”. If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure you of complete continuity. 2. When an image on the film is obliterated with a round black mark it is an indication that the film inspector noticed either blurred copy because of movement during exposure, or duplicate copy.