The Genetic Constituent of Varicose Vein Pathogenesis As a Key for Future Treatment Option Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

IDENTIFICATION of CELL SURFACE MARKERS WHICH CORRELATE with SALL4 in a B-CELL ACUTE LYMPHOBLASTIC LEUKEMIA with T(8;14)
IDENTIFICATION of CELL SURFACE MARKERS WHICH CORRELATE WITH SALL4 in a B-CELL ACUTE LYMPHOBLASTIC LEUKEMIA WITH T(8;14) DISCOVERED THROUGH BIOINFORMATIC ANALYSIS of MICROARRAY GENE EXPRESSION DATA The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:38962442 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ,'(17,),&$7,21 2) &(// 685)$&( 0$5.(56 :+,&+ &255(/$7( :,7+ 6$// ,1 $ %&(// $&87( /<03+2%/$67,& /(8.(0,$ :,7+ W ',6&29(5(' 7+528*+ %,2,1)250$7,& $1$/<6,6 2) 0,&52$55$< *(1( (;35(66,21 '$7$ 52%(57 3$8/ :(,1%(5* $ 7KHVLV 6XEPLWWHG WR WKH )DFXOW\ RI 7KH +DUYDUG 0HGLFDO 6FKRRO LQ 3DUWLDO )XOILOOPHQW RI WKH 5HTXLUHPHQWV IRU WKH 'HJUHH RI 0DVWHU RI 0HGLFDO 6FLHQFHV LQ ,PPXQRORJ\ +DUYDUG 8QLYHUVLW\ %RVWRQ 0DVVDFKXVHWWV -XQH Thesis Advisor: Dr. Li Chai Author: Robert Paul Weinberg Department of Pathology Candidate MMSc in Immunology Brigham and Womens’ Hospital Harvard Medical School 77 Francis Street 25 Shattuck Street Boston, MA 02215 Boston, MA 02215 IDENTIFICATION OF CELL SURFACE MARKERS WHICH CORRELATE WITH SALL4 IN A B-CELL ACUTE LYMPHOBLASTIC LEUKEMIA WITH TRANSLOCATION t(8;14) DISCOVERED THROUGH BIOINFORMATICS ANALYSIS OF MICROARRAY GENE EXPRESSION DATA Abstract Acute Lymphoblastic Leukemia (ALL) is the most common leukemia in children, causing signficant morbidity and mortality annually in the U.S. -

PARSANA-DISSERTATION-2020.Pdf
DECIPHERING TRANSCRIPTIONAL PATTERNS OF GENE REGULATION: A COMPUTATIONAL APPROACH by Princy Parsana A dissertation submitted to The Johns Hopkins University in conformity with the requirements for the degree of Doctor of Philosophy Baltimore, Maryland July, 2020 © 2020 Princy Parsana All rights reserved Abstract With rapid advancements in sequencing technology, we now have the ability to sequence the entire human genome, and to quantify expression of tens of thousands of genes from hundreds of individuals. This provides an extraordinary opportunity to learn phenotype relevant genomic patterns that can improve our understanding of molecular and cellular processes underlying a trait. The high dimensional nature of genomic data presents a range of computational and statistical challenges. This dissertation presents a compilation of projects that were driven by the motivation to efficiently capture gene regulatory patterns in the human transcriptome, while addressing statistical and computational challenges that accompany this data. We attempt to address two major difficulties in this domain: a) artifacts and noise in transcriptomic data, andb) limited statistical power. First, we present our work on investigating the effect of artifactual variation in gene expression data and its impact on trans-eQTL discovery. Here we performed an in-depth analysis of diverse pre-recorded covariates and latent confounders to understand their contribution to heterogeneity in gene expression measurements. Next, we discovered 673 trans-eQTLs across 16 human tissues using v6 data from the Genotype Tissue Expression (GTEx) project. Finally, we characterized two trait-associated trans-eQTLs; one in Skeletal Muscle and another in Thyroid. Second, we present a principal component based residualization method to correct gene expression measurements prior to reconstruction of co-expression networks. -

Pharmacy and Poisons (Third and Fourth Schedule Amendment) Order 2017
Q UO N T FA R U T A F E BERMUDA PHARMACY AND POISONS (THIRD AND FOURTH SCHEDULE AMENDMENT) ORDER 2017 BR 111 / 2017 The Minister responsible for health, in exercise of the power conferred by section 48A(1) of the Pharmacy and Poisons Act 1979, makes the following Order: Citation 1 This Order may be cited as the Pharmacy and Poisons (Third and Fourth Schedule Amendment) Order 2017. Repeals and replaces the Third and Fourth Schedule of the Pharmacy and Poisons Act 1979 2 The Third and Fourth Schedules to the Pharmacy and Poisons Act 1979 are repealed and replaced with— “THIRD SCHEDULE (Sections 25(6); 27(1))) DRUGS OBTAINABLE ONLY ON PRESCRIPTION EXCEPT WHERE SPECIFIED IN THE FOURTH SCHEDULE (PART I AND PART II) Note: The following annotations used in this Schedule have the following meanings: md (maximum dose) i.e. the maximum quantity of the substance contained in the amount of a medicinal product which is recommended to be taken or administered at any one time. 1 PHARMACY AND POISONS (THIRD AND FOURTH SCHEDULE AMENDMENT) ORDER 2017 mdd (maximum daily dose) i.e. the maximum quantity of the substance that is contained in the amount of a medicinal product which is recommended to be taken or administered in any period of 24 hours. mg milligram ms (maximum strength) i.e. either or, if so specified, both of the following: (a) the maximum quantity of the substance by weight or volume that is contained in the dosage unit of a medicinal product; or (b) the maximum percentage of the substance contained in a medicinal product calculated in terms of w/w, w/v, v/w, or v/v, as appropriate. -
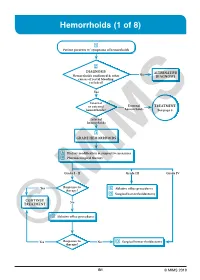
Hemorrhoids (1 of 8)
Hemorrhoids (1 of 8) 1 Patient presents w/ symptoms of hemorrhoids 2 DIAGNOSIS No ALTERNATIVE Hemorrhoids confi rmed & other DIAGNOSIS causes of rectal bleeding excluded? Yes Internal or external External TREATMENT hemorrhoids? hemorrhoids See page 3 Internal hemorrhoids 3 GRADE HEMORRHOIDS A Dietary modifi cation & supportive measures B Pharmacological therapy Grade I - II Grade III Grade IV Yes Response to C Ablative offi ce procedures therapy? D Surgical hemorrhoidectomy CONTINUE No TREATMENT C Ablative offi ce procedures Yes Response to No D Surgical hemorrhoidectomy ©therapy? MIMS B1 © MIMS 2019 Hemorrhoids (2 of 8) 1 SYMPTOMS ATTRIBUTED TO HEMORRHOIDS • Rectal bleeding - Most common presenting symptom - Bright red blood which may drip or squirt into the toilet bowl or scanty amounts may be seen on toilet tissue • Discomfort due to rectal protrusion or lump • Anal pain • HEMORRHOIDS Anal itching 2 DIAGNOSIS Medical History • Assess nature, duration & severity of symptoms - Ask about bleeding, its amount & frequency - Ask about presence of prolapsing tissue, its timing & reproducibility • Elicit possible risk factors for development of hemorrhoidal symptoms - Low-fi ber diets cause small-caliber stools, resulting in straining during defecation & engorgement of hemorrhoids - Prolonged sitting on a toilet which may cause a problem in the venous return in the perianal area - Pregnancy - Advanced age • Th e signs & symptoms of hemorrhoids are not specifi c to the disease, so care must be taken to avoid missing other causes of pathology • Obtain -

Early Growth Response 1 Regulates Hematopoietic Support and Proliferation in Human Primary Bone Marrow Stromal Cells
Hematopoiesis SUPPLEMENTARY APPENDIX Early growth response 1 regulates hematopoietic support and proliferation in human primary bone marrow stromal cells Hongzhe Li, 1,2 Hooi-Ching Lim, 1,2 Dimitra Zacharaki, 1,2 Xiaojie Xian, 2,3 Keane J.G. Kenswil, 4 Sandro Bräunig, 1,2 Marc H.G.P. Raaijmakers, 4 Niels-Bjarne Woods, 2,3 Jenny Hansson, 1,2 and Stefan Scheding 1,2,5 1Division of Molecular Hematology, Department of Laboratory Medicine, Lund University, Lund, Sweden; 2Lund Stem Cell Center, Depart - ment of Laboratory Medicine, Lund University, Lund, Sweden; 3Division of Molecular Medicine and Gene Therapy, Department of Labora - tory Medicine, Lund University, Lund, Sweden; 4Department of Hematology, Erasmus MC Cancer Institute, Rotterdam, the Netherlands and 5Department of Hematology, Skåne University Hospital Lund, Skåne, Sweden ©2020 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2019.216648 Received: January 14, 2019. Accepted: July 19, 2019. Pre-published: August 1, 2019. Correspondence: STEFAN SCHEDING - [email protected] Li et al.: Supplemental data 1. Supplemental Materials and Methods BM-MNC isolation Bone marrow mononuclear cells (BM-MNC) from BM aspiration samples were isolated by density gradient centrifugation (LSM 1077 Lymphocyte, PAA, Pasching, Austria) either with or without prior incubation with RosetteSep Human Mesenchymal Stem Cell Enrichment Cocktail (STEMCELL Technologies, Vancouver, Canada) for lineage depletion (CD3, CD14, CD19, CD38, CD66b, glycophorin A). BM-MNCs from fetal long bones and adult hip bones were isolated as reported previously 1 by gently crushing bones (femora, tibiae, fibulae, humeri, radii and ulna) in PBS+0.5% FCS subsequent passing of the cell suspension through a 40-µm filter. -

Discovery of 318 New Risk Loci for Type 2 Diabetes and Related Vascular Outcomes Among 1.4 Million Participants in a Multi-Ancestry Meta-Analysis
ARTICLES https://doi.org/10.1038/s41588-020-0637-y Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis Marijana Vujkovic 1,2,112, Jacob M. Keaton3,4,5,6,112, Julie A. Lynch 7,8, Donald R. Miller9,10, Jin Zhou 11,12, Catherine Tcheandjieu13,14,15, Jennifer E. Huffman 16, Themistocles L. Assimes 13,14, Kimberly Lorenz 1,17,18, Xiang Zhu 13,19, Austin T. Hilliard 13,14, Renae L. Judy1,20, Jie Huang16,21, Kyung M. Lee 7, Derek Klarin 16,22,23,24, Saiju Pyarajan16,25,26, John Danesh27, Olle Melander28, Asif Rasheed29, Nadeem H. Mallick 30, Shahid Hameed30, Irshad H. Qureshi31,32, Muhammad Naeem Afzal31,32, Uzma Malik31,32, Anjum Jalal33, Shahid Abbas33, Xin Sheng 2, Long Gao17, Klaus H. Kaestner17, Katalin Susztak 2, Yan V. Sun 34,35, Scott L. DuVall 7,36, Kelly Cho16,25, Jennifer S. Lee13,14, J. Michael Gaziano16,25, Lawrence S. Phillips 34,37, James B. Meigs23,26,38, Peter D. Reaven 11,39, Peter W. Wilson34,40, Todd L. Edwards 4,41, Daniel J. Rader 2,17, Scott M. Damrauer 1,20, Christopher J. O’Donnell 16,25,26, Philip S. Tsao 13,14, The HPAP Consortium*, Regeneron Genetics Center*, VA Million Veteran Program*, Kyong-Mi Chang 1,2,42,112, Benjamin F. Voight 1,17,18,112 ✉ and Danish Saleheen 29,43,44,112 ✉ We investigated type 2 diabetes (T2D) genetic susceptibility via multi-ancestry meta-analysis of 228,499 cases and 1,178,783 controls in the Million Veteran Program (MVP), DIAMANTE, Biobank Japan and other studies. -

Pharmaceutical Appendix to the Tariff Schedule 2
Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2007) (Rev. 2) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACIDUM LIDADRONICUM 63132-38-7 ABAFUNGIN 129639-79-8 ACIDUM SALCAPROZICUM 183990-46-7 ABAMECTIN 65195-55-3 ACIDUM SALCLOBUZICUM 387825-03-8 ABANOQUIL 90402-40-7 ACIFRAN 72420-38-3 ABAPERIDONUM 183849-43-6 ACIPIMOX 51037-30-0 ABARELIX 183552-38-7 ACITAZANOLAST 114607-46-4 ABATACEPTUM 332348-12-6 ACITEMATE 101197-99-3 ABCIXIMAB 143653-53-6 ACITRETIN 55079-83-9 ABECARNIL 111841-85-1 ACIVICIN 42228-92-2 ABETIMUSUM 167362-48-3 ACLANTATE 39633-62-0 ABIRATERONE 154229-19-3 ACLARUBICIN 57576-44-0 ABITESARTAN 137882-98-5 ACLATONIUM NAPADISILATE 55077-30-0 ABLUKAST 96566-25-5 ACODAZOLE 79152-85-5 ABRINEURINUM 178535-93-8 ACOLBIFENUM 182167-02-8 ABUNIDAZOLE 91017-58-2 ACONIAZIDE 13410-86-1 ACADESINE 2627-69-2 ACOTIAMIDUM 185106-16-5 ACAMPROSATE 77337-76-9 -

Vr Meds Ex01 3B 0825S Coding Manual Supplement Page 1
vr_meds_ex01_3b_0825s Coding Manual Supplement MEDNAME OTHER_CODE ATC_CODE SYSTEM THER_GP PHRM_GP CHEM_GP SODIUM FLUORIDE A12CD01 A01AA01 A A01 A01A A01AA SODIUM MONOFLUOROPHOSPHATE A12CD02 A01AA02 A A01 A01A A01AA HYDROGEN PEROXIDE D08AX01 A01AB02 A A01 A01A A01AB HYDROGEN PEROXIDE S02AA06 A01AB02 A A01 A01A A01AB CHLORHEXIDINE B05CA02 A01AB03 A A01 A01A A01AB CHLORHEXIDINE D08AC02 A01AB03 A A01 A01A A01AB CHLORHEXIDINE D09AA12 A01AB03 A A01 A01A A01AB CHLORHEXIDINE R02AA05 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S01AX09 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S02AA09 A01AB03 A A01 A01A A01AB CHLORHEXIDINE S03AA04 A01AB03 A A01 A01A A01AB AMPHOTERICIN B A07AA07 A01AB04 A A01 A01A A01AB AMPHOTERICIN B G01AA03 A01AB04 A A01 A01A A01AB AMPHOTERICIN B J02AA01 A01AB04 A A01 A01A A01AB POLYNOXYLIN D01AE05 A01AB05 A A01 A01A A01AB OXYQUINOLINE D08AH03 A01AB07 A A01 A01A A01AB OXYQUINOLINE G01AC30 A01AB07 A A01 A01A A01AB OXYQUINOLINE R02AA14 A01AB07 A A01 A01A A01AB NEOMYCIN A07AA01 A01AB08 A A01 A01A A01AB NEOMYCIN B05CA09 A01AB08 A A01 A01A A01AB NEOMYCIN D06AX04 A01AB08 A A01 A01A A01AB NEOMYCIN J01GB05 A01AB08 A A01 A01A A01AB NEOMYCIN R02AB01 A01AB08 A A01 A01A A01AB NEOMYCIN S01AA03 A01AB08 A A01 A01A A01AB NEOMYCIN S02AA07 A01AB08 A A01 A01A A01AB NEOMYCIN S03AA01 A01AB08 A A01 A01A A01AB MICONAZOLE A07AC01 A01AB09 A A01 A01A A01AB MICONAZOLE D01AC02 A01AB09 A A01 A01A A01AB MICONAZOLE G01AF04 A01AB09 A A01 A01A A01AB MICONAZOLE J02AB01 A01AB09 A A01 A01A A01AB MICONAZOLE S02AA13 A01AB09 A A01 A01A A01AB NATAMYCIN A07AA03 A01AB10 A A01 -

Protein-DNA Interactions of Pul34, an Essential Human Cytomegalovirus DNA
Protein-DNA Interactions of pUL34, an Essential Human Cytomegalovirus DNA- Binding Protein A dissertation presented to the faculty of the College of Arts and Sciences of Ohio University In partial fulfillment of the requirements for the degree Doctor of Philosophy Mark D. Slayton August 2018 © 2018 Mark D. Slayton. All Rights Reserved. 2 This dissertation titled Protein-DNA Interactions of pUL34, an Essential Human Cytomegalovirus DNA- Binding Protein by MARK D. SLAYTON has been approved for the Department of Molecular and Cellular Biology and the College of Arts and Sciences by Bonita J. Biegalke Associate Professor of Biomedical Sciences Joseph Shields Interim Dean, College of Arts and Sciences 3 ABSTRACT SLAYTON, MARK D., Ph.D., August 2018, Molecular and Cellular Biology Protein-DNA Interactions of pUL34, an Essential Human Cytomegalovirus DNA- Binding Protein Director of Dissertation: Bonita J. Biegalke Human cytomegalovirus (HCMV) is primarily an opportunistic pathogen in human, causing significant disease in immunocompromised individuals. A large, double- stranded DNA genome (~230 kilobases) provides the coding capacity for over 200 genes, of which only 25% are required for viral replication in cell culture. The viral UL34 gene encodes sequence-specific DNA-binding proteins (pUL34) which are essential for replication, and viruses lacking the proper expression of pUL34 cannot replicate in cell culture. Interactions of pUL34 with DNA binding sites (US3 and US9?) represses transcription of (these) two viral immune evasion genes that are dispensable for replication in cell culture. There are 12 additional predicted pUL34-binding sites present in the HCMV genome (strain AD169), with three of them concentrated near the HCMV origin of lytic replication (oriLyt). -

Deciphering the Molecular Profile of Plaques, Memory Decline And
ORIGINAL RESEARCH ARTICLE published: 16 April 2014 AGING NEUROSCIENCE doi: 10.3389/fnagi.2014.00075 Deciphering the molecular profile of plaques, memory decline and neuron loss in two mouse models for Alzheimer’s disease by deep sequencing Yvonne Bouter 1†,Tim Kacprowski 2,3†, Robert Weissmann4, Katharina Dietrich1, Henning Borgers 1, Andreas Brauß1, Christian Sperling 4, Oliver Wirths 1, Mario Albrecht 2,5, Lars R. Jensen4, Andreas W. Kuss 4* andThomas A. Bayer 1* 1 Division of Molecular Psychiatry, Georg-August-University Goettingen, University Medicine Goettingen, Goettingen, Germany 2 Department of Bioinformatics, Institute of Biometrics and Medical Informatics, University Medicine Greifswald, Greifswald, Germany 3 Department of Functional Genomics, Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, Germany 4 Human Molecular Genetics, Department for Human Genetics of the Institute for Genetics and Functional Genomics, Institute for Human Genetics, University Medicine Greifswald, Ernst-Moritz-Arndt University Greifswald, Greifswald, Germany 5 Institute for Knowledge Discovery, Graz University of Technology, Graz, Austria Edited by: One of the central research questions on the etiology of Alzheimer’s disease (AD) is the Isidro Ferrer, University of Barcelona, elucidation of the molecular signatures triggered by the amyloid cascade of pathological Spain events. Next-generation sequencing allows the identification of genes involved in disease Reviewed by: Isidro Ferrer, University of Barcelona, processes in an unbiased manner. We have combined this technique with the analysis of Spain two AD mouse models: (1) The 5XFAD model develops early plaque formation, intraneu- Dietmar R. Thal, University of Ulm, ronal Ab aggregation, neuron loss, and behavioral deficits. (2)TheTg4–42 model expresses Germany N-truncated Ab4–42 and develops neuron loss and behavioral deficits albeit without plaque *Correspondence: formation. -

Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DIX to the HTSUS—Continued
20558 Federal Register / Vol. 60, No. 80 / Wednesday, April 26, 1995 / Notices DEPARMENT OF THE TREASURY Services, U.S. Customs Service, 1301 TABLE 1.ÐPHARMACEUTICAL APPEN- Constitution Avenue NW, Washington, DIX TO THE HTSUSÐContinued Customs Service D.C. 20229 at (202) 927±1060. CAS No. Pharmaceutical [T.D. 95±33] Dated: April 14, 1995. 52±78±8 ..................... NORETHANDROLONE. A. W. Tennant, 52±86±8 ..................... HALOPERIDOL. Pharmaceutical Tables 1 and 3 of the Director, Office of Laboratories and Scientific 52±88±0 ..................... ATROPINE METHONITRATE. HTSUS 52±90±4 ..................... CYSTEINE. Services. 53±03±2 ..................... PREDNISONE. 53±06±5 ..................... CORTISONE. AGENCY: Customs Service, Department TABLE 1.ÐPHARMACEUTICAL 53±10±1 ..................... HYDROXYDIONE SODIUM SUCCI- of the Treasury. NATE. APPENDIX TO THE HTSUS 53±16±7 ..................... ESTRONE. ACTION: Listing of the products found in 53±18±9 ..................... BIETASERPINE. Table 1 and Table 3 of the CAS No. Pharmaceutical 53±19±0 ..................... MITOTANE. 53±31±6 ..................... MEDIBAZINE. Pharmaceutical Appendix to the N/A ............................. ACTAGARDIN. 53±33±8 ..................... PARAMETHASONE. Harmonized Tariff Schedule of the N/A ............................. ARDACIN. 53±34±9 ..................... FLUPREDNISOLONE. N/A ............................. BICIROMAB. 53±39±4 ..................... OXANDROLONE. United States of America in Chemical N/A ............................. CELUCLORAL. 53±43±0 -

Multiple Aberrations of Chromosome 3P Detected in Oral Premalignant Lesions
Cancer Prevention Research Multiple Aberrations of Chromosome 3p Detected in Oral Premalignant Lesions Ivy F.L. Tsui,1,3 Miriam P. Rosin,2 Lewei Zhang,4 Raymond T. Ng5 and Wan L. Lam1,3 Abstract The study of oral premalignant lesions (OPL) is crucial to the identification of initiating genetic events in oral cancer. However, these lesions are minute in size, making it a challenge to recover sufficient DNA from microdissected cells for comprehensive genomic analysis. As a step toward identifying genetic aberrations associated with oral cancer progression, we used tiling-path array comparative genomic hybridization to compare alterations on chromosome 3p for 71 OPLs against 23 oral squamous cell carcinomas. 3p was chosen because although it is frequently altered in oral cancers and has been associated with progression risk, its alteration status has only been evaluated at a small number of loci in OPLs. We identified six recurrent losses in this region that were shared between high-grade dysplasias and oral squamous cell carcinomas, including a 2.89-Mbp deletion spanning the FHIT gene (previously implicated in oral cancer progression). When the alteration status for these six regions was examined in 24 low-grade dysplasias with known progression outcome, we observed that they occurred at a significantly higher fre- quency in low-grade dysplasias that later progressed to later-stage disease (P < 0.003). Moreover, parallel analysis of all profiled tissues showed that the extent of overall genomic alteration at 3p increased with histologic stage. This first high-resolution analysis of chromo- some arm 3p in OPLs represents a significant step toward predicting progression risk in early preinvasive disease and provides a keen example of how genomic instability escalates with progression to invasive cancer.