Quantitative Trait Locus Mapping Reveals an Independent Genetic Basis for Joint Divergence in Leaf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Lineage-Specific Mapping of Quantitative Trait Loci
Heredity (2013) 111, 106–113 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Lineage-specific mapping of quantitative trait loci C Chen1 and K Ritland We present an approach for quantitative trait locus (QTL) mapping, termed as ‘lineage-specific QTL mapping’, for inferring allelic changes of QTL evolution along with branches in a phylogeny. We describe and analyze the simplest case: by adding a third taxon into the normal procedure of QTL mapping between pairs of taxa, such inferences can be made along lineages to a presumed common ancestor. Although comparisons of QTL maps among species can identify homology of QTLs by apparent co- location, lineage-specific mapping of QTL can classify homology into (1) orthology (shared origin of QTL) versus (2) paralogy (independent origin of QTL within resolution of map distance). In this light, we present a graphical method that identifies six modes of QTL evolution in a three taxon comparison. We then apply our model to map lineage-specific QTLs for inbreeding among three taxa of yellow monkey-flower: Mimulus guttatus and two inbreeders M. platycalyx and M. micranthus, but critically assuming outcrossing was the ancestral state. The two most common modes of homology across traits were orthologous (shared ancestry of mutation for QTL alleles). The outbreeder M. guttatus had the fewest lineage-specific QTL, in accordance with the presumed ancestry of outbreeding. Extensions of lineage-specific QTL mapping to other types of data and crosses, and to inference of ancestral QTL state, are discussed. Heredity (2013) 111, 106–113; doi:10.1038/hdy.2013.24; published online 24 April 2013 Keywords: Quantitative trait locus mapping; evolution of mating systems; Mimulus INTRODUCTION common ancestor of these two taxa, back to the common ancestor of Knowledge of the genetic architecture of complex traits can offer all three taxa and forwards again to the third taxon. -
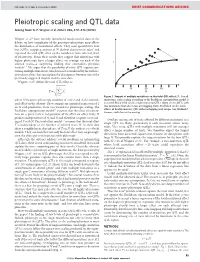
Pleiotropic Scaling and QTL Data Arising From: G
NATURE | Vol 456 | 4 December 2008 BRIEF COMMUNICATIONS ARISING Pleiotropic scaling and QTL data Arising from: G. P. Wagner et al. Nature 452, 470–473 (2008) Wagner et al.1 have recently introduced much-needed data to the debate on how complexity of the genotype–phenotype map affects the distribution of mutational effects. They used quantitative trait loci (QTLs) mapping analysis of 70 skeletal characters in mice2 and regressed the total QTL effect on the number of traits affected (level of pleiotropy). From their results they suggest that mutations with higher pleiotropy have a larger effect, on average, on each of the T affected traits—a surprising finding that contradicts previous models3–7. We argue that the possibility of some QTL regions con- taining multiple mutations, which was not considered by the authors, introduces a bias that can explain the discrepancy between one of the previously suggested models and the new data. Wagner et al.1 define the total QTL effect as vffiffiffiffiffiffiffiffiffiffiffiffiffi u uXN 10 20 30 ~t 2 T Ai N i~1 Figure 1 | Impact of multiple mutations on the total QTL effect, T. For all where N measures pleiotropy (number of traits) and Ai the normal- mutations, strict scaling according to the Euclidean superposition model is ized effect on the ith trait. They compare an empirical regression of T assumed. Black filled circles, single-mutation QTLs. Open circles, QTLs with on N with predictions from two models for pleiotropic scaling. The two mutations that affect non-overlapping traits. Red filled circles, total Euclidean superposition model3,4 assumes that the effect of a muta- effects of double-mutant QTL with overlapping trait ranges (see Methods). -

1 ROBERT PLOMIN on Behavioral Genetics Genetic Links Common
Genetic links • Background – Why has educational psychology ben so slow ROBERT PLOMIN to accept genetics? on Behavioral Genetics • Beyond nature vs. nurture – 3 findings from genetic research with far- reaching implications for education: October, 2004 The nature of nurture The abnormal is normal Generalist genes • Molecular Genetics Quantitative genetics: twin and adoption studies Common medical disorders Identical twins Fraternal twins (monozygotic, MZ) (dizygotic, DZ) 1 Twin concordances in TEDS at 7 years Twins Early Development Study (TEDS) for low reading and maths Perceptions of nature/nurture: % parents (N=1340) and teachers Beyond nature vs. nurture: (N=667) indicating that genes are at least as important as environment (Walker & Plomin, Ed. Psych., in press) The nature of nurture • Genetic influence on experience • Genotype-environment correlation – Children’s genetic propensities are correlated with their experiences passive evocative active 2 Beyond nature vs. nurture: Genotype-environment correlation The nature of nurture • Passive • Genotype-environment correlation – e.g., family background (SES) and school – Children’s genetic propensities are correlated with achievement their experiences passive • Evocative evocative – Children evoke experiences for genetic reasons Active • Active • Molecular genetics – Children actively create environments that foster – Find genes associated with learning experiences their genetic propensities Beyond nature vs. nurture: The one-gene one-disability (OGOD) model The abnormal is normal • Common learning disabilities are the quantitative extreme of the same genetic and environmental factors responsible for normal variation in learning abilities. • There are no common learning disabilities, just the extremes of normal variation. 3 OGOD (rare) and QTL (common) disorders The quantitative trait locus (QTL) model Beyond nature vs. -

Epigenome-Wide Study Identified Methylation Sites Associated With
nutrients Article Epigenome-Wide Study Identified Methylation Sites Associated with the Risk of Obesity Majid Nikpay 1,*, Sepehr Ravati 2, Robert Dent 3 and Ruth McPherson 1,4,* 1 Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, 40 Ruskin St–H4208, Ottawa, ON K1Y 4W7, Canada 2 Plastenor Technologies Company, Montreal, QC H2P 2G4, Canada; [email protected] 3 Department of Medicine, Division of Endocrinology, University of Ottawa, the Ottawa Hospital, Ottawa, ON K1Y 4E9, Canada; [email protected] 4 Atherogenomics Laboratory, University of Ottawa Heart Institute, Ottawa, ON K1Y 4W7, Canada * Correspondence: [email protected] (M.N.); [email protected] (R.M.) Abstract: Here, we performed a genome-wide search for methylation sites that contribute to the risk of obesity. We integrated methylation quantitative trait locus (mQTL) data with BMI GWAS information through a SNP-based multiomics approach to identify genomic regions where mQTLs for a methylation site co-localize with obesity risk SNPs. We then tested whether the identified site contributed to BMI through Mendelian randomization. We identified multiple methylation sites causally contributing to the risk of obesity. We validated these findings through a replication stage. By integrating expression quantitative trait locus (eQTL) data, we noted that lower methylation at cg21178254 site upstream of CCNL1 contributes to obesity by increasing the expression of this gene. Higher methylation at cg02814054 increases the risk of obesity by lowering the expression of MAST3, whereas lower methylation at cg06028605 contributes to obesity by decreasing the expression of SLC5A11. Finally, we noted that rare variants within 2p23.3 impact obesity by making the cg01884057 Citation: Nikpay, M.; Ravati, S.; Dent, R.; McPherson, R. -

QUANTITATIVE TRAIT LOCUS ANALYSIS: in Several Variants (I.E., Alleles)
TECHNOLOGIES FROM THE FIELD QUANTITATIVE TRAIT LOCUS ANALYSIS: in several variants (i.e., alleles). QTL analysis allows one MULTIPLE CROSS AND HETEROGENEOUS to map, with some precision, the genomic position of STOCK MAPPING these alleles. For many researchers in the alcohol field, the break Robert Hitzemann, Ph.D.; John K. Belknap, Ph.D.; through with respect to the genetic determination of alcohol- and Shannon K. McWeeney, Ph.D. related behaviors occurred when Plomin and colleagues (1991) made the seminal observation that a specific panel of RI mice (i.e., the BXD panel) could be used to identify KEY WORDS: Genetic theory of alcohol and other drug use; genetic the physical location of (i.e., to map) QTLs for behavioral factors; environmental factors; behavioral phenotype; behavioral phenotypes. Because the phenotypes of the different strains trait; quantitative trait gene (QTG); inbred animal strains; in this panel had been determined for many alcohol- recombinant inbred (RI) mouse strains; quantitative traits; related traits, researchers could readily apply the strategy quantitative trait locus (QTL) mapping; multiple cross mapping of RI–QTL mapping (Gora-Maslak et al. 1991). Although (MCM); heterogeneous stock (HS) mapping; microsatellite investigators recognized early on that this panel was not mapping; animal models extensive enough to answer all questions, the emerging data illustrated the rich genetic complexity of alcohol- related phenotypes (Belknap 1992; Plomin and McClearn ntil well into the 1990s, both preclinical and clinical 1993). research focused on finding “the” gene for human The next advance came with the development of diseases, including alcoholism. This focus was rein U microsatellite maps (Dietrich et al. -

THE JEPSON GLOBE a Newsletter from the Friends of the Jepson Herbarium
THE JEPSON GLOBE A Newsletter from the Friends of The Jepson Herbarium VOLUME 29 NUMBER 1, Spring 2019 Curator’s column: Don Kyhos’s Upcoming changes in the Con- legacy in California botany sortium of California Herbaria By Bruce G. Baldwin By Jason Alexander In early April, my Ph.D. advisor, In January, the Northern California Donald W. Kyhos (UC Davis) turns 90, Botanists Association hosted their 9th fittingly during one of the California Botanical Symposium in Chico, Cali- desert’s most spectacular blooms in fornia. The Consortium of California recent years. Don’s many contributions Herbaria (CCH) was invited to present to desert botany and plant evolution on upcoming changes. The CCH be- in general are well worth celebrating gan as a data aggregator for California here for their critical importance to our vascular plant specimen data and that understanding of the California flora. remains its primary purpose to date. Those old enough to have used Munz’s From 2003 until 2017, the CCH grew A California Flora may recall seeing in size to over 2.2 million specimen re- the abundant references to Raven and cords from 36 institutions. Responding Kyhos’s chromosome numbers, which to requests from participants to display reflect a partnership between Don and specimen data from all groups of plants Peter Raven that yielded a tremendous Rudi Schmid at Antelope Valley Califor- and fungi, from all locations (including body of cytogenetic information about nia Poppy Reserve on 7 April 2003. Photo those outside California), we have de- our native plants. Don’s talents as a by Ray Cranfill. -

Article the Power to Detect Quantitative Trait Loci Using
The Power to Detect Quantitative Trait Loci Using Resequenced, Experimentally Evolved Populations of Diploid, Sexual Organisms James G. Baldwin-Brown,*,1 Anthony D. Long,1 and Kevin R. Thornton1 1Department of Ecology and Evolutionary Biology, University of California, Irvine *Corresponding author: E-mail: [email protected]. Associate editor: John Parsch Abstract A novel approach for dissecting complex traits is to experimentally evolve laboratory populations under a controlled environment shift, resequence the resulting populations, and identify single nucleotide polymorphisms (SNPs) and/or genomic regions highly diverged in allele frequency. To better understand the power and localization ability of such an evolve and resequence (E&R) approach, we carried out forward-in-time population genetics simulations of 1 Mb genomic regions under a large combination of experimental conditions, then attempted to detect significantly diverged SNPs. Our analysis indicates that the ability to detect differentiation between populations is primarily affected by selection coef- ficient, population size, number of replicate populations, and number of founding haplotypes. We estimate that E&R studies can detect and localize causative sites with 80% success or greater when the number of founder haplotypes is over 500, experimental populations are replicated at least 25-fold, population size is at least 1,000 diploid individuals, and the selection coefficient on the locus of interest is at least 0.1. More achievable experimental designs (less replicated, fewer founder haplotypes, smaller effective population size, and smaller selection coefficients) can have power of greater than 50% to identify a handful of SNPs of which one is likely causative. Similarly, in cases where s 0.2, less demanding experimental designs can yield high power. -

Glossary/Index
Glossary 03/08/2004 9:58 AM Page 119 GLOSSARY/INDEX The numbers after each term represent the chapter in which it first appears. additive 2 allele 2 When an allele’s contribution to the variation in a One of two or more alternative forms of a gene; a single phenotype is separately measurable; the independent allele for each gene is inherited separately from each effects of alleles “add up.” Antonym of nonadditive. parent. ADHD/ADD 6 Alzheimer’s disease 5 Attention Deficit Hyperactivity Disorder/Attention A medical disorder causing the loss of memory, rea- Deficit Disorder. Neurobehavioral disorders character- soning, and language abilities. Protein residues called ized by an attention span or ability to concentrate that is plaques and tangles build up and interfere with brain less than expected for a person's age. With ADHD, there function. This disorder usually first appears in persons also is age-inappropriate hyperactivity, impulsive over age sixty-five. Compare to early-onset Alzheimer’s. behavior or lack of inhibition. There are several types of ADHD: a predominantly inattentive subtype, a predomi- amino acids 2 nantly hyperactive-impulsive subtype, and a combined Molecules that are combined to form proteins. subtype. The condition can be cognitive alone or both The sequence of amino acids in a protein, and hence pro- cognitive and behavioral. tein function, is determined by the genetic code. adoption study 4 amnesia 5 A type of research focused on families that include one Loss of memory, temporary or permanent, that can result or more children raised by persons other than their from brain injury, illness, or trauma. -

A Latitudinal Cline in Physiology Is Associated with More Variable Precipitation in Erythranthe Cardinalis
bioRxiv preprint doi: https://doi.org/10.1101/080952; this version posted September 3, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Grow with the flow: a latitudinal cline in physiology is associated with more variable precipitation in Erythranthe cardinalis Christopher D. Muir1;∗ and Amy L. Angert1;2 1 Biodiversity Research Centre and Department of Botany, University of British Columbia, Vancouver, BC, Canada 2 Department of Zoology, University of British Columbia, Vancouver, BC, Canada ∗corresponding author: Chris Muir, [email protected] Running Head: Latitudinal cline and climate in Erythranthe Key words: local adaptation, cline, photosynthesis, growth rate, monkeyflower, Mimulus Data are currently available on GitHub and will be archived on Dryad upon publication. Acknowledgements Erin Warkman and Lisa Lin helped collect data. CDM was supported by a Biodiversity Postdoctoral Fellowship funded by the NSERC CREATE program. ALA was supported by an NSERC Discovery Grant and a grant from the National Science Foundation (DEB 0950171). Four anonymous referees provided constructive comments on earlier versions of this manuscript. 1 bioRxiv preprint doi: https://doi.org/10.1101/080952; this version posted September 3, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Grow with the flow: a latitudinal cline in physiology is associated with more variable precipitation in Erythranthe cardinalis Abstract Local adaptation is commonly observed in nature: organisms perform well in their natal environment, but poorly outside it. -

Vascular Plants of Santa Cruz County, California
ANNOTATED CHECKLIST of the VASCULAR PLANTS of SANTA CRUZ COUNTY, CALIFORNIA SECOND EDITION Dylan Neubauer Artwork by Tim Hyland & Maps by Ben Pease CALIFORNIA NATIVE PLANT SOCIETY, SANTA CRUZ COUNTY CHAPTER Copyright © 2013 by Dylan Neubauer All rights reserved. No part of this publication may be reproduced without written permission from the author. Design & Production by Dylan Neubauer Artwork by Tim Hyland Maps by Ben Pease, Pease Press Cartography (peasepress.com) Cover photos (Eschscholzia californica & Big Willow Gulch, Swanton) by Dylan Neubauer California Native Plant Society Santa Cruz County Chapter P.O. Box 1622 Santa Cruz, CA 95061 To order, please go to www.cruzcps.org For other correspondence, write to Dylan Neubauer [email protected] ISBN: 978-0-615-85493-9 Printed on recycled paper by Community Printers, Santa Cruz, CA For Tim Forsell, who appreciates the tiny ones ... Nobody sees a flower, really— it is so small— we haven’t time, and to see takes time, like to have a friend takes time. —GEORGIA O’KEEFFE CONTENTS ~ u Acknowledgments / 1 u Santa Cruz County Map / 2–3 u Introduction / 4 u Checklist Conventions / 8 u Floristic Regions Map / 12 u Checklist Format, Checklist Symbols, & Region Codes / 13 u Checklist Lycophytes / 14 Ferns / 14 Gymnosperms / 15 Nymphaeales / 16 Magnoliids / 16 Ceratophyllales / 16 Eudicots / 16 Monocots / 61 u Appendices 1. Listed Taxa / 76 2. Endemic Taxa / 78 3. Taxa Extirpated in County / 79 4. Taxa Not Currently Recognized / 80 5. Undescribed Taxa / 82 6. Most Invasive Non-native Taxa / 83 7. Rejected Taxa / 84 8. Notes / 86 u References / 152 u Index to Families & Genera / 154 u Floristic Regions Map with USGS Quad Overlay / 166 “True science teaches, above all, to doubt and be ignorant.” —MIGUEL DE UNAMUNO 1 ~ACKNOWLEDGMENTS ~ ANY THANKS TO THE GENEROUS DONORS without whom this publication would not M have been possible—and to the numerous individuals, organizations, insti- tutions, and agencies that so willingly gave of their time and expertise. -

Mapping Quantitative Trait Loci in Selected Breeding Populations: a Segregation Distortion Approach
Heredity (2015) 115, 538–546 & 2015 Macmillan Publishers Limited All rights reserved 0018-067X/15 www.nature.com/hdy ORIGINAL ARTICLE Mapping quantitative trait loci in selected breeding populations: A segregation distortion approach Y Cui1,2, F Zhang1,JXu1,3,ZLi1 and S Xu2 Quantitative trait locus (QTL) mapping is often conducted in line-crossing experiments where a sample of individuals is randomly selected from a pool of all potential progeny. QTLs detected from such an experiment are important for us to understand the genetic mechanisms governing a complex trait, but may not be directly relevant to plant breeding if they are not detected from the breeding population where selection is targeting for. QTLs segregating in one population may not necessarily segregate in another population. To facilitate marker-assisted selection, QTLs must be detected from the very population which the selection is targeting. However, selected breeding populations often have depleted genetic variation with small population sizes, resulting in low power in detecting useful QTLs. On the other hand, if selection is effective, loci controlling the selected trait will deviate from the expected Mendelian segregation ratio. In this study, we proposed to detect QTLs in selected breeding populations via the detection of marker segregation distortion in either a single population or multiple populations using the same selection scheme. Simulation studies showed that QTL can be detected in strong selected populations with selected population sizes as small as 25 plants. We applied the new method to detect QTLs in two breeding populations of rice selected for high grain yield. Seven QTLs were identified, four of which have been validated in advanced generations in a follow-up study. -

A Taxonomic Conspectus of Phrymaceae: a Narrowed Circumscriptions for Mimulus , New and Resurrected Genera, and New Names and Combinations
Barker, W.R., G.L. Nesom, P.M. Beardsley, and N.S. Fraga. 2012. A taxonomic conspectus of Phrymaceae: A narrowed circumscriptions for Mimulus , new and resurrected genera, and new names and combinations. Phytoneuron 2012-39: 1–60. Published 16 May 2012. ISSN 2153 733X A TAXONOMIC CONSPECTUS OF PHRYMACEAE: A NARROWED CIRCUMSCRIPTION FOR MIMULUS, NEW AND RESURRECTED GENERA, AND NEW NAMES AND COMBINATIONS W.R. (B ILL ) BARKER State Herbarium of South Australia, Kent Town SA 5071, AUSTRALIA; and Australian Centre for Evolutionary Biology and Biodiversity University of Adelaide Adelaide SA 5000, AUSTRALIA [email protected] GUY L. NESOM 2925 Hartwood Drive Fort Worth, Texas 76109, USA [email protected] PAUL M. BEARDSLEY Biological Sciences Department California State Polytechnic University Pomona, California 91768, USA [email protected] NAOMI S. FRAGA Rancho Santa Ana Botanic Garden Claremont, California 91711-3157, USA [email protected] ABSTRACT A revised taxonomic classification of Phrymaceae down to species level is presented, based on molecular-phylogenetic and morpho-taxonomic studies, setting a framework for ongoing work. In the concept adopted, the family includes 188 species divided into 13 genera. All species as currently understood are listed and assigned to genera and sections which in several cases have new circumscriptions requiring many new combinations. Four main clades are recognized. Clade A. An Asian-African-Australasian-centered clade of 7 genera: Mimulus L. sensu stricto (7 species) of North America, Asia to Africa, and Australasia is sister to an Australian-centered group that comprises Elacholoma (2 species), Glossostigma (5 species), Microcarpaea (2 species), Peplidium (4 species), Uvedalia (2 species) and a new monotypic genus Thyridia , described here.