Supplementary Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chimeric Gpcrs Mimic Distinct Signaling Pathways and Modulate Microglia Responses
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.21.449162; this version posted June 21, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Chimeric GPCRs mimic distinct signaling pathways and modulate microglia responses Rouven Schulz, Medina Korkut-Demirbaş, Gloria Colombo, Sandra Siegert† Author affiliation: Institute of Science and Technology (IST) Austria, Am Campus 1, 3400 Klosterneuburg, Austria † Corresponding author: [email protected] Keywords: G protein coupled receptor (GPCR), DREADD, β2-adrenergic receptor (β2AR/ADRB2), microglia, inflammation, GPR65, GPR109A/HCAR2 Abstract G protein-coupled receptors (GPCRs) regulate multiple processes ranging from cell growth and immune responses to neuronal signal transmission. However, ligands for many GPCRs remain unknown, suffer from off-target effects or have poor bioavailability. Additional challenges exist to dissect cell type-specific responses when the same GPCR is expressed on different cells within the body. Here, we overcome these limitations by engineering DREADD-based GPCR chimeras that selectively bind their agonist clozapine-N-oxide (CNO) and mimic a GPCR-of-interest. We show that the chimeric DREADD-β2-adrenergic receptor (β2AR/ADRB2) triggers comparable responses to levalbuterol on second messenger and kinase activity, post-translational modifications, and protein-protein interactions. Moreover, we successfully recapitulate β2AR-mediated filopodia formation in microglia, a β2AR-expressing immune cell that can drive inflammation in the nervous system. To further dissect microglial inflammation, we compared DREADD-β2AR with two additionally designed DREADD-based chimeras mimicking GPR65 and GPR109A/HCAR2, both enriched in microglia. -

G Protein-Coupled Receptors As New Therapeutic Targets for Type 2 Diabetes
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Diabetologia (2016) 59:229–233 DOI 10.1007/s00125-015-3825-z MINI-REVIEW G protein-coupled receptors as new therapeutic targets for type 2 diabetes Frank Reimann1 & Fiona M. Gribble 1 Received: 31 October 2015 /Accepted: 9 November 2015 /Published online: 12 December 2015 # The Author(s) 2015. This article is published with open access at Springerlink.com Abstract G protein-coupled receptors (GPCRs) in the gut– GLP1R Glucagon-like peptide 1 receptor brain–pancreatic axis are key players in the postprandial con- GPBAR1 G protein-coupled bile acid receptor trol of metabolism and food intake. A number of intestinally GPCR G protein-coupled receptor located receptors have been implicated in the chemo-detection of ingested nutrients, and in the pancreatic islets and nervous system GPCRs play essential roles in the detection of many Therapeutics that promote insulin secretion have been a main- hormones and neurotransmitters. Because of the diversity, stay of type 2 diabetes treatment for many years. However, cell-specific expression and ‘druggability’ of the GPCR su- with the rising impact of obesity on the incidence of type 2 perfamily, these receptors are popular targets for therapeutic diabetes comes an increasing need to target body weight as development. This review will outline current and potential well as blood glucose control. Recent years have witnessed an future approaches to develop GPCR agonists for the treatment increasing interest in the gut endocrine system as a source of of type 2 diabetes. -

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

A Guide to Glutamate Receptors
A guide to glutamate receptors 1 Contents Glutamate receptors . 4 Ionotropic glutamate receptors . 4 - Structure ........................................................................................................... 4 - Function ............................................................................................................ 5 - AMPA receptors ................................................................................................. 6 - NMDA receptors ................................................................................................. 6 - Kainate receptors ............................................................................................... 6 Metabotropic glutamate receptors . 8 - Structure ........................................................................................................... 8 - Function ............................................................................................................ 9 - Group I: mGlu1 and mGlu5. .9 - Group II: mGlu2 and mGlu3 ................................................................................. 10 - Group III: mGlu4, mGlu6, mGlu7 and mGlu8 ............................................................ 10 Protocols and webinars . 11 - Protocols ......................................................................................................... 11 - Webinars ......................................................................................................... 12 References and further reading . 13 Excitatory synapse pathway -

Trace Amine-Associated Receptor 1 Activation Regulates Glucose-Dependent
Trace amine-associated receptor 1 activation regulates glucose-dependent insulin secretion in pancreatic beta cells in vitro by ©Arun Kumar A thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree of Master of Science Department of Biochemistry, Faculty of Science Memorial University of Newfoundland FEBRUARY 2021 St. John’s, Newfoundland and Labrador i Abstract Trace amines are a group of endogenous monoamines which exert their action through a family of G protein-coupled receptors known as trace amine-associated receptors (TAARs). TAAR1 has been reported to regulate insulin secretion from pancreatic beta cells in vitro and in vivo. This study investigates the mechanism(s) by which TAAR1 regulates insulin secretion. The insulin secreting rat INS-1E -cell line was used for the study. Cells were pre-starved (30 minutes) and then incubated with varying concentrations of glucose (2.5 – 20 mM) or KCl (3.6 – 60 mM) for 2 hours in the absence or presence of various concentrations of the selective TAAR1 agonist RO5256390. Secreted insulin per well was quantified using ELISA and normalized to the total protein content of individual cultures. RO5256390 significantly (P < 0.0001) increased glucose- stimulated insulin secretion in a dose-dependent manner, with no effect on KCl-stimulated insulin secretion. Affymetrix-microarray data analysis identified genes (Gnas, Gng7, Gngt1, Gria2, Cacna1e, Kcnj8, and Kcnj11) whose expression was associated with changes in TAAR1 in response to changes in insulin secretion in pancreatic beta cell function. The identified potential links to TAAR1 supports the regulation of glucose-stimulated insulin secretion through KATP ion channels. -
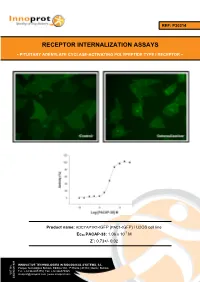
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Gastrointestinal Defense Mechanisms
REVIEW CURRENT OPINION Gastrointestinal defense mechanisms Hyder Said a,b and Jonathan D. Kaunitzb,c Purpose of review To summarize and illuminate the recent findings regarding gastroduodenal mucosal defense mechanisms and the specific biomolecules involved in regulating this process, such as glucagon-like peptides (GLPs). Recent findings There has been a growing interest in luminal nutrient chemosensing and its physiological effects throughout the digestive system. From the ingestion of food in the oral cavity to the processing and absorption of nutrients in the intestines, nutrient chemosensing receptors signal the production and release of numerous bioactive peptides from enteroendocrine cells, such as the proglucagon-derived peptides. There has been a major emphasis on two proglucagon-derived peptides, namely GLP-1 and GLP-2, due to their apparent beneficial effect on gut structure, function, and on metabolic processes. As an incretin, GLP-1 not only enhances the effect and release of insulin on pancreatic bcells but also has been implicated in having trophic effects on the intestinal epithelium. In addition, GLP-2, the other major proglucagon-derived peptide, has potent intestinotrophic effects, such as increasing the rate of mucosal stem cell proliferation, mucosal blood flow, and fluid absorption, as well as augmenting the rate of duodenal bicarbonate secretion to improve gastric mucosal health and longevity. Summary Understanding the mechanisms underlying nutrient chemosensing and how it relates to GLP release can further elucidate how the gut functions in response to cellular changes and disturbances. Furthermore, a more in-depth comprehension of GLP release and its tissue-specific effects will help improve the utility of GLP-1 and GLP-2 receptor agonists in clinical settings. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Lysophosphatidic Acid Signaling in the Nervous System
Neuron Review Lysophosphatidic Acid Signaling in the Nervous System Yun C. Yung,1,3 Nicole C. Stoddard,1,2,3 Hope Mirendil,1 and Jerold Chun1,* 1Molecular and Cellular Neuroscience Department, Dorris Neuroscience Center, The Scripps Research Institute, La Jolla, CA 92037, USA 2Biomedical Sciences Graduate Program, University of California, San Diego School of Medicine, La Jolla, CA 92037, USA 3Co-first author *Correspondence: [email protected] http://dx.doi.org/10.1016/j.neuron.2015.01.009 The brain is composed of many lipids with varied forms that serve not only as structural components but also as essential signaling molecules. Lysophosphatidic acid (LPA) is an important bioactive lipid species that is part of the lysophospholipid (LP) family. LPA is primarily derived from membrane phospholipids and signals through six cognate G protein-coupled receptors (GPCRs), LPA1-6. These receptors are expressed on most cell types within central and peripheral nervous tissues and have been functionally linked to many neural pro- cesses and pathways. This Review covers a current understanding of LPA signaling in the nervous system, with particular focus on the relevance of LPA to both physiological and diseased states. Introduction LPA synthesis/degradative enzymes (reviewed in Sigal et al., The human brain is composed of approximately 60%–70% lipids 2005; Brindley and Pilquil, 2009; Perrakis and Moolenaar, by dry weight (Svennerholm et al., 1994). These lipids can be 2014). In view of the broad neurobiological influences of LPA divided into two major pools, structural and signaling, which signaling, its dysregulation may lead to diverse neuropathologies include well-known families such as cholesterol, fatty acids, ei- (Bandoh et al., 2000; Houben and Moolenaar, 2011; Yung et al., cosanoids, endocannabinoids, and prostaglandins (Figure 1). -

Interplay Between Gating and Block of Ligand-Gated Ion Channels
brain sciences Review Interplay between Gating and Block of Ligand-Gated Ion Channels Matthew B. Phillips 1,2, Aparna Nigam 1 and Jon W. Johnson 1,2,* 1 Department of Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA; [email protected] (M.B.P.); [email protected] (A.N.) 2 Center for Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA * Correspondence: [email protected]; Tel.: +1-(412)-624-4295 Received: 27 October 2020; Accepted: 26 November 2020; Published: 1 December 2020 Abstract: Drugs that inhibit ion channel function by binding in the channel and preventing current flow, known as channel blockers, can be used as powerful tools for analysis of channel properties. Channel blockers are used to probe both the sophisticated structure and basic biophysical properties of ion channels. Gating, the mechanism that controls the opening and closing of ion channels, can be profoundly influenced by channel blocking drugs. Channel block and gating are reciprocally connected; gating controls access of channel blockers to their binding sites, and channel-blocking drugs can have profound and diverse effects on the rates of gating transitions and on the stability of channel open and closed states. This review synthesizes knowledge of the inherent intertwining of block and gating of excitatory ligand-gated ion channels, with a focus on the utility of channel blockers as analytic probes of ionotropic glutamate receptor channel function. Keywords: ligand-gated ion channel; channel block; channel gating; nicotinic acetylcholine receptor; ionotropic glutamate receptor; AMPA receptor; kainate receptor; NMDA receptor 1. Introduction Neuronal information processing depends on the distribution and properties of the ion channels found in neuronal membranes. -

Metabolite Sensing Gpcrs: Promising Therapeutic Targets for Cancer Treatment?
cells Review Metabolite Sensing GPCRs: Promising Therapeutic Targets for Cancer Treatment? Jesús Cosín-Roger 1,*, Dolores Ortiz-Masia 2 , Maria Dolores Barrachina 3 and Sara Calatayud 3 1 Hospital Dr. Peset, Fundación para la Investigación Sanitaria y Biomédica de la Comunitat Valenciana, FISABIO, 46017 Valencia, Spain 2 Departament of Medicine, Faculty of Medicine, University of Valencia, 46010 Valencia, Spain; [email protected] 3 Departament of Pharmacology and CIBER, Faculty of Medicine, University of Valencia, 46010 Valencia, Spain; [email protected] (M.D.B.); [email protected] (S.C.) * Correspondence: [email protected]; Tel.: +34-963851234 Received: 30 September 2020; Accepted: 21 October 2020; Published: 23 October 2020 Abstract: G-protein-coupled receptors constitute the most diverse and largest receptor family in the human genome, with approximately 800 different members identified. Given the well-known metabolic alterations in cancer development, we will focus specifically in the 19 G-protein-coupled receptors (GPCRs), which can be selectively activated by metabolites. These metabolite sensing GPCRs control crucial processes, such as cell proliferation, differentiation, migration, and survival after their activation. In the present review, we will describe the main functions of these metabolite sensing GPCRs and shed light on the benefits of their potential use as possible pharmacological targets for cancer treatment. Keywords: G-protein-coupled receptor; metabolite sensing GPCR; cancer 1. Introduction G-protein-coupled receptors (GPCRs) are characterized by a seven-transmembrane configuration, constitute the largest and most ubiquitous family of plasma membrane receptors, and regulate virtually all known physiological processes in humans [1,2]. This family includes almost one thousand genes that were initially classified on the basis of sequence homology into six classes (A–F), where classes D and E were not found in vertebrates [3]. -

Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Tohoku J. Exp. Med., 2011,Differential 223, 161-176 Gene Expression in OPCs, Oligodendrocytes and Type II Astrocytes 161 Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes Jian-Guo Hu,1,2,* Yan-Xia Wang,3,* Jian-Sheng Zhou,2 Chang-Jie Chen,4 Feng-Chao Wang,1 Xing-Wu Li1 and He-Zuo Lü1,2 1Department of Clinical Laboratory Science, The First Affiliated Hospital of Bengbu Medical College, Bengbu, P.R. China 2Anhui Key Laboratory of Tissue Transplantation, Bengbu Medical College, Bengbu, P.R. China 3Department of Neurobiology, Shanghai Jiaotong University School of Medicine, Shanghai, P.R. China 4Department of Laboratory Medicine, Bengbu Medical College, Bengbu, P.R. China Oligodendrocyte precursor cells (OPCs) are bipotential progenitor cells that can differentiate into myelin-forming oligodendrocytes or functionally undetermined type II astrocytes. Transplantation of OPCs is an attractive therapy for demyelinating diseases. However, due to their bipotential differentiation potential, the majority of OPCs differentiate into astrocytes at transplanted sites. It is therefore important to understand the molecular mechanisms that regulate the transition from OPCs to oligodendrocytes or astrocytes. In this study, we isolated OPCs from the spinal cords of rat embryos (16 days old) and induced them to differentiate into oligodendrocytes or type II astrocytes in the absence or presence of 10% fetal bovine serum, respectively. RNAs were extracted from each cell population and hybridized to GeneChip with 28,700 rat genes. Using the criterion of fold change > 4 in the expression level, we identified 83 genes that were up-regulated and 89 genes that were down-regulated in oligodendrocytes, and 92 genes that were up-regulated and 86 that were down-regulated in type II astrocytes compared with OPCs.