The Partial Mitochondrial Sequence of the Old World Stingless Bee, Tetragonula Pagdeni (2013) Journal of Genetics, Pp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Test Using Drosophila Melanogaster
bioRxiv preprint doi: https://doi.org/10.1101/553123; this version posted February 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Does Long-Term Selection for Development Time Result in Canalization: A Test Using Drosophila melanogaster * * Shampa M. Ghosh1,2 , K. M. Satish1,3, J. Mohan1,4 and Amitabh Joshi1 Author affiliations 1: Evolutionary Biology Laboratory, Evolutionary & Organismal Biology Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Jakkur, Bengaluru 560064, India 2: Present address: Fly Research Laboratory, School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT) Campus 11, Bhubaneswar 751024, India 3: Present address: Department of Biotechnology, College of Agriculture, Navile, University of Agricultural and Horticultural Sciences, Shivamogga 577201, India 4: Present address: Centre of Excellence in Genomics and Translational Medicine, University of Tartu, Tartu 50411, Estonia *Correspondence: Dr. Shampa M. Ghosh: [email protected] Dr. Amitabh Joshi: [email protected] Abstract Canalization denotes the robustness of a trait against genetic or environmental perturbation. Plasticity, in contrast indicates the environmental sensitivity of a trait. Stabilizing selection is thought to increase canalization of a trait, whereas directional selection is often thought to lead to decanalization. However, the relationship between selection, canalization and plasticity remains largely unclear. Experimental evolution is a powerful approach for addressing fundamental questions in evolution. Here, we ask whether long-term directional selection for reduced pre-adult development time in Drosophila melanogaster results in the evolution of increased canalization for development time, the trait under primary selection. -
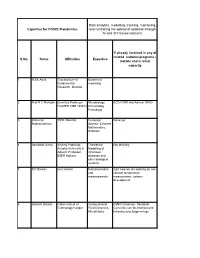
Data Analytics and Modeling
Data analytics, modelling, tracking, monitoring, Expertise for COVID /Pandemics and controlling the spread of outbreak through AI and IOT-based solutions If already involved in any of related national programs : S.No. Name Affiliation Expertise Details and in what capacity 1 H. M. Antia Tata Institute of Numerical Fundamental modelling Research, Mumbai 2 Prof R C Mahajan Emeritus Professor Microbiology ECD ICMR and Advisor WHO PGIMER CHD 160012 Immunology Parasitogy 3 Jaikumar TIFR, Mumbai Computer None yet. Radhakrishnan Science, Discrete Mathematics, Software 4 Somdatta Sinha Visiting Professor, Theoretical Not officially. Ashoka University & Modelling of Adjunct Professor, infectious IISER Kolkata diseases and other biological systems 5 S K Biswas iiser mohali Instrumentation right now we are working on non and contact temperature measurements measurement system development 6 Gautam Biswas Indian Intitute of Computational GIAN (Chairman, Sectional Technology Kanpur Fluid Dynamics, Committee on Mechanical and Microfluidics Infrastructure Engineering) 7 Ram Ramaswamy IIT Delhi Modeling - 8 Sandip Paul Ramanujan Fellow NGS data NA analysis, adaptive evolution 9 Amish kumar Ph.D. Student Bioinformatics After submission of my Ph.D thesis I am working as visitor research student at Department of plant sciences under Project entitled as "TIGR2ESS: Transforming India's Green Revolution by Research and Empowerment for Sustainable food Supplies". 10 Surajit Assistant Professor, Immunology, None Bhattacharjee Department of Infection Biology Molecular -

Year : 2015-16
LIST OF RTI Nodal Officers Year : 2015-16 Sr.No Ministry/Department/Organisation Nodal Officer Contact Address Phone No/Fax/ Email Name/Designation 1 Cabinet Secretariat Shri Sunil Mishra Rashtrapati Bhawan Director & CPIO New Delhi 1 Cabinet Secretariat 110001 23018467 2 Department of Atomic Energy Shri P Ramakrishnan Central Office, Western Chief Administrator Anushaktinagar, 1 Atomic Energy Education Society Mumbai - 400 094 02225565049 [email protected] Dr. Pankaj Tandon Niyamak Bhavan, Scientific Officer F Anushaktinagar, 2 Atomic Energy Regulatory Board Mumbai- 400 094 25990659 [email protected] Shri M.B. Verma Atomic Minerals Atomic Minerals Directorate for Additional Director (Op 1-10-153/156, AMD 3 Exploration and Research Hyderabad - 500 016 04027766472 [email protected] Dr. P.M. Satya Sai Head, Commissioning 04427480044 Scientific Officer (H) Nuclear Recycle Group 4427480252 4 BARC Facilities, Kalpakkam Kalpakkam, [email protected] Shri Sanjay Pradhan TNRPO, NRB, BARC, 02525244299 Chief Superitendent Ghivali Post, Palghar 2525244158 5 Bhabha Atomic Resarch Centre (Tarapur) Pin 401502 [email protected] Shri. B. P. Joshi Central Complex, 3rd 02225505330 Chief Administrative Trombay, 2225505151 6 Bhabha Atomic Research Centre Mumbai - 400 085 [email protected] Smt. K. Malathi BHAVINI 04427480915 Bharatiya Nabhikiya Vidhyut Nigam Ltd. Senior Manager (HR) Kalpakkam - 603 102 4427480640 7 (BHAVINI) Kancheepuram Dist. [email protected] Shri M.C. Dinakaran BRIT, Project House, 02225573534 Board of Radiation and Isotope General Manager (PIO) Anushakti Nagar 2225562161 8 Technology Mumbai- 400 094. [email protected] Deputy Secretary Anushakti Bhavan 22839961 Deputy Secretary CSM Marg, 22048476 9 Department of Atomic Energy Mumbai-400001 [email protected] Shri V.K. -
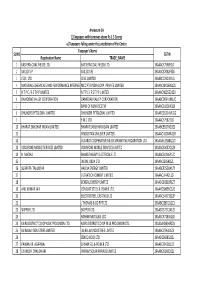
FINAL DISTRIBUTION.Xlsx
Annexure-1A 1)Taxpayers with turnover above Rs 1.5 Crores a) Taxpayers falling under the jurisdiction of the Centre Taxpayer's Name SL NO GSTIN Registration Name TRADE_NAME 1 EASTERN COAL FIELDS LTD. EASTERN COAL FIELDS LTD. 19AAACE7590E1ZI 2 SAIL (D.S.P) SAIL (D.S.P) 19AAACS7062F6Z6 3 CESC LTD. CESC LIMITED 19AABCC2903N1ZL 4 MATERIALS CHEMICALS AND PERFORMANCE INTERMEDIARIESMCC PTA PRIVATE INDIA CORP.LIMITED PRIVATE LIMITED 19AAACM9169K1ZU 5 N T P C / F S T P P LIMITED N T P C / F S T P P LIMITED 19AAACN0255D1ZV 6 DAMODAR VALLEY CORPORATION DAMODAR VALLEY CORPORATION 19AABCD0541M1ZO 7 BANK OF NOVA SCOTIA 19AAACB1536H1ZX 8 DHUNSERI PETGLOBAL LIMITED DHUNSERI PETGLOBAL LIMITED 19AAFCD5214M1ZG 9 E M C LTD 19AAACE7582J1Z7 10 BHARAT SANCHAR NIGAM LIMITED BHARAT SANCHAR NIGAM LIMITED 19AABCB5576G3ZG 11 HINDUSTAN UNILEVER LIMITED 19AAACH1004N1ZR 12 GUJARAT COOPERATIVE MILKS MARKETING FEDARATION LTD 19AAAAG5588Q1ZT 13 VODAFONE MOBILE SERVICES LIMITED VODAFONE MOBILE SERVICES LIMITED 19AAACS4457Q1ZN 14 N MADHU BHARAT HEAVY ELECTRICALS LTD 19AAACB4146P1ZC 15 JINDAL INDIA LTD 19AAACJ2054J1ZL 16 SUBRATA TALUKDAR HALDIA ENERGY LIMITED 19AABCR2530A1ZY 17 ULTRATECH CEMENT LIMITED 19AAACL6442L1Z7 18 BENGAL ENERGY LIMITED 19AADCB1581F1ZT 19 ANIL KUMAR JAIN CONCAST STEEL & POWER LTD.. 19AAHCS8656C1Z0 20 ELECTROSTEEL CASTINGS LTD 19AAACE4975B1ZP 21 J THOMAS & CO PVT LTD 19AABCJ2851Q1Z1 22 SKIPPER LTD. SKIPPER LTD. 19AADCS7272A1ZE 23 RASHMI METALIKS LTD 19AACCR7183E1Z6 24 KAIRA DISTRICT CO-OP MILK PRO.UNION LTD. KAIRA DISTRICT CO-OP MILK PRO.UNION LTD. 19AAAAK8694F2Z6 25 JAI BALAJI INDUSTRIES LIMITED JAI BALAJI INDUSTRIES LIMITED 19AAACJ7961J1Z3 26 SENCO GOLD LTD. 19AADCS6985J1ZL 27 PAWAN KR. AGARWAL SHYAM SEL & POWER LTD. 19AAECS9421J1ZZ 28 GYANESH CHAUDHARY VIKRAM SOLAR PRIVATE LIMITED 19AABCI5168D1ZL 29 KARUNA MANAGEMENT SERVICES LIMITED 19AABCK1666L1Z7 30 SHIVANANDAN TOSHNIWAL AMBUJA CEMENTS LIMITED 19AAACG0569P1Z4 31 SHALIMAR HATCHERIES LIMITED SHALIMAR HATCHERIES LTD 19AADCS6537J1ZX 32 FIDDLE IRON & STEEL PVT. -

Basic Research : Its Role In
Published by: The National Academy of Sciences, India (NASI) Year of Publication: 2018 For further information, please contact: The National Academy of Sciences, India (NASI) 5, Lajpat Rai Road, Allahabad, Uttar Pradesh-211002 Phone: +91 (0532) 2640224, Fax: +91 (532) 2641183 Email: [email protected]; [email protected] Website: www.nasi.ac.in PROCEEDINGS OF THE SYMPOSIUM ON BASIC RESEARCH – IT’S ROLE IN NATIONAL DEVELOPMENT 87TH ANNUAL SESSION OF NASI 8TH -10TH DEC 2017 Venue:- Savitribai Phule Pune University, Ganeshkhind Road, Pune CONTENTS FOREWORD PREFACE I. INAUGURAL SESSION Welcome Address 3 Prof. Nitin R. Karmalkar, Vice-Chancellor, Savitribai Phule Pune University About the Pune Chapter 5 Dr. Dilip Dattaray Dhavale, Chairman, NASI Local Chapter About the Annual Symposium 7 Dr. Manju Sharma (Convener) Distinguished Woman Scientist Chair, NASI and Former Secretary, Department of Biotechnology, Government of India Role of Space Research in National Development 9 Dr. Kiran Kumar (Guest of Honour) Secretary, Department of Space, Chairman, Space Commission and Chairman, ISRO Presidential Address 18 Dr. Anil Kakodkar, President, NASI Inaugural Address 20 Shri C. H. Vidyasagar Rao, Governor of Maharashtra Vote of Thanks 24 Prof. Veena Tandon, General Secretary, NASI Game Changing in Indian Science, Technology and Innovation 26 Dr. R. A. Mashelkar, FRS (Chief Guest) National Research Professor Theme Publication: Basic Research as an Integral Component of a Self Reliant 33 Base of Science and Technology (Its Role, Relevance, Support, Areas of Thrust) Late Prof. M.G.K. Menon, FRS Invited Article: Basic Science: A Clinician’s Perception 64 Prof. P.N. Tandon, National Research Professor and Former President, NASI II. -

Contemporary Excitement in New Biology NAGALAND UNIVERSITY
UGC Sponsored National Conference on Contemporary Excitement in New Biology National Advisors (CENB- 2018) Prof. SN. Hegde, Mysuru October 30-31, 2018 Prof. M. Venkateswarlu, Shimoga Prof. LS. Shashidhara, Pune Organized by Prof. Amitabh Joshi, Bangalore Dr. T. Madhan Mohan, New Delhi Dr. Kumarasamy Thangaraj Hyderabad Prof. Pawan Sharma, New Delhi Prof. Jagat Kumar Roy, Varanasi Prof. RNS. Yadav, Dibrugarh NAGALAND UNIVERSITY Prof. Ramesh Sharma, Shillong Prof. S. Rama Rao, Shillong (A Central University) Prof. Prabodh Bora, Guwahati Prof. Prasad Burra, Haryana DEPARTMENT OF ZOOLOGY Prof. DK. Sharma, Guwahati Headquarters: LUMAMI, ZUNHEBOTO DISTRICT- Dr. Longvah, Hyderabad 798627 NAGALAND, INDIA Prof. Neelu Nangia, Bangalore Prof. Kamal Jaiswal, Lucknow Prof. Kaushal, Nainital Organizing Committee Prof. OP. Singh, Shillong Prof. BBP. Gupta, Shillong Patron Prof. SB. Prasad, Shillong Prof. Pardeshi Lal Vice Chancellor, Nagaland University Convener(s) Prof. SU. Ahmed Prof. LN. Kakati Department of Zoology, Nagaland Confirmed Speakers: University Prof. Anupam Chatterjee (NEHU, Shillong, Organizing Secretary Meghalaya (Keynote speaker) Prof. Sarat Chandra Yenisetti Prof. BK. Konwar (Tejpur University, Head, Department of Zoology, Nagaland Assam) University Prof. KRS. Sambasiva Rao (Mizoram University, Mizoram) Treasurer Prof. JC. Kalita (Guwahati University, Dr. Bendang Ao Assam) Department of Zoology, Nagaland Prof. Jharna Chakraborty (Rajiv Gandhi University University, Arunachal Pradesh) Prof. AK. Mukherjee (Tejpur University, Members -

Indian National Science Academy Bahadur Shah Zafar Marg, New Delhi - 110002
INDIAN NATIONAL SCIENCE ACADEMY BAHADUR SHAH ZAFAR MARG, NEW DELHI - 110002 Minutes of the General Body Meeting of the Indian National Science Academy held on 27 July, 2017 in the Academy premises. The following Fellows were present: Professor Ajay K Sood, President, INSA Professor Anurag Sharma, Vice-President (Fellowship Affairs) Professor Kankan Bhattacharyya, Vice-President (Science Promotion) Professor NR Jagannathan, Vice-President (Resource Management) Professor SC Lakhotia, Vice-President (Informatics and Publications) Professor LS Shashidhara, Vice-President (Science & Society) Professor Faizan Ahmad Professor Talat Ahmad Dr AC Banerjea Professor Subhasis Chaudhuri Dr Madhu Dikshit Dr Kunal Ghosh Dr Swapan K Ghosh Professor LC Gupta Professor Amitabh Joshi Dr Sushil Kumar Professor Nibir Mandal Professor NK Mehra Professor HY Mohan Ram Dr Ganesh Pandey Professor G Parthasarathy Professor IBS Passi Professor Sarva Jit Singh Professor Somdatta Sinha Professor PN Tandon Dr KC Upadhyaya Professor K Veluthambi President, INSA welcomed all the Fellows to the General body meeting. Thereafter, the regular agenda items were taken up. 1. Condolence at the passing away of the following distinguished Fellows: The sad demise of Professors Bhola Nath Dhawan, Arumugam Gnanam, Udupi Ramachandra Rao, Arun Kumar Sharma and Yash Pal distinguished Fellows of the Academy was reported. The obituary notes were read by the President and all those present stood in silence for a minute as a mark of respect to the deceased. 2. Confirmation of minutes of Ordinary General Meeting held on 26 April, 2017. The minutes of the Ordinary General Meeting held on 26 April, 2017 were read by Professor Anurag Sharma, Vice-President. -

Contemporary Issues in Evolutionary Biology
Journal of Genetics, Vol. 96, No. 3, July 2017, pp. 399–400 DOI 10.1007/s12041-017-0806-7 Contemporary issues in evolutionary biology Preface We are delighted to bring to the readers, a set of peer-reviewed papers on evolutionary biology, published as a special issue of the Journal of Genetics. These papers emanated from ruminations upon and discussions at the Foundations of Evolutionary Theory: the Ongoing Synthesis meeting at Coorg, India, in February 2014, and the Foundations of Biology meetings at Pune, India, in April 2015 and March 2016. One of us (LSS), along with Sutirth Dey (IISER Pune) and Amitabh Joshi (JNCASR, Bengaluru), organized these meetings in order to bring together a diverse set of interested academics to discuss and argue about fundamental issues in the subject. These discussions included, among others, the possible consequences of nonDNA-based inheritance—epigenetics and cultural evolution, niche construction, and developmental mechanisms on our understanding of the evolutionary process, speciation, complexity in biology, and constructing a formal evolutionary theory. This special issue begins with an original article by K. P. Mohanan on the Conceptual Foundations of Evolution- ary Thought, in which evolution is conceptualized as symmetry breaking, thereby attempting to integrate physical, biological (macroevolution), and cultural (societal) evolution. Biological evolution and cultural evolution are fur- ther examined conceptually as the emergence and persistence of traits rather than taxa. Structural and functional constraints are invoked to explain the contrast between the emergence of novelty and the resistance to novelty (per- sistence). The second article presents a review of various species concepts, and thoughts about species concepts versus the delimitation of species. -

The Year Book 2019
THE YEAR BOOK 2019 INDIAN ACADEMY OF SCIENCES Bengaluru Postal Address: Indian Academy of Sciences Post Box No. 8005 C.V. Raman Avenue Sadashivanagar Post, Raman Research Institute Campus Bengaluru 560 080 India Telephone : +91-80-2266 1200, +91-80-2266 1203 Fax : +91-80-2361 6094 Email : [email protected], [email protected] Website : www.ias.ac.in © 2019 Indian Academy of Sciences Information in this Year Book is updated up to 22 February 2019. Editorial & Production Team: Nalini, B.R. Thirumalai, N. Vanitha, M. Venugopal, M.S. Published by: Executive Secretary, Indian Academy of Sciences Text formatted by WINTECS Typesetters, Bengaluru (Ph. +91-80-2332 7311) Printed by Lotus Printers Pvt. Ltd., Bengaluru CONTENTS Page Section A: Indian Academy of Sciences Memorandum of Association ................................................... 2 Role of the Academy ............................................................... 4 Statutes .................................................................................. 7 Council for the period 2019–2021 ............................................ 18 Office Bearers ......................................................................... 19 Former Presidents ................................................................... 20 Activities – a profile ................................................................. 21 Academy Document on Scientific Values ................................. 25 The Academy Trust ................................................................. 33 Section B: Professorships -

Jawharalal Nehru Annual Rep-2011-12
ISSN.0973-9319 ANNUAL REPORT 2011-2012 JAWAHARLAL NEHRU CENTRE FOR ADVANCED SCIENTIFIC RESEARCH (A Deemed to be University) Jakkur, Bangalore – 560 064. Website: http://www.jncasr.ac.in CONTENTS Page No. The Centre Foreword ........................................................................................................................................... 1 Introduction .......................................................................................................................................... 3 Objectives ........................................................................................................................................... 4 Progress ........................................................................................................................................... 5 Highlights of research and other activities....................................................................................... 7 Activities Chart ..................................................................................................................................... 12 Organisation Chart .............................................................................................................................. 13 The Organisation Council of Management ...................................................................................................................... 14 Finance Committee .............................................................................................................................. -

Female Asian Elephants in Nagarahole National Park, Southern India ” Has Been Carried out by Mr
RESOURCE AVAILABILITY , WITHIN -CLAN AND BETWEEN -CLAN AGONISTIC INTERACTIONS , AND DOMINANCE RELATIONSHIPS AMONGST FEMALE ASIAN ELEPHANTS IN NAGARAHOLE NATIONAL PARK , SOUTHERN INDIA A thesis submitted for the degree of Doctor of Philosophy by Hansraj Gautam Evolutionary and Organismal Biology Unit Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru 560064, India. September 2019 ii CERTIFICATE This is to certify that the work presented in this thesis titled “ Resource Availability, Within-Clan and Between-Clan Agonistic Interactions, and Dominance Relationships amongst Female Asian Elephants in Nagarahole National Park, Southern India ” has been carried out by Mr. Hansraj Gautam under my supervision at the Evolutionary and Organismal Biology Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru, India, and that the results in this thesis have not previously formed the basis for the award of any other degree, diploma, or fellowship. Date: Prof. T.N.C. Vidya iii iv DECLARATION I declare that the matter presented in my thesis titled “Resource Availability, Within-Clan and Between-Clan Agonistic Interactions, and Dominance Relationships amongst Female Asian Elephants in Nagarahole National Park, Southern India ” is the result of studies carried out by me at the Evolutionary and Organismal Biology Unit of the Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore, India, under the supervision of Prof. T.N.C. Vidya, and that this work has not been submitted elsewhere for any other degree. In keeping with the general practice of reporting scientific observations, due acknowledgement has been made wherever the work described has been based on the findings of other investigators. Any omission, which might have occurred by oversight, is regretted. -

Activity Report (2013-2015)
2 0 1 3 ICTS— 2 0 1 5 REPORTTATA INSTITUTE OF FUNDAMENTAL RESEARCH International Centre for Theoretical Sciences (ICTS-TIFR) Shivakote, Hesaraghatta Hobli, Bengaluru North 560 089 www.icts.res.in ii ICTS REPORT 2013-15 | DIRECTOR’S REPORT ICTS REPORT 2013-15 | DIRECTOR’S REPORT iii CONTENTS 1) DIRECTOR’S REPORT PAGE 1 2) RESEARCH REPORTS PAGE 7 3) ASSOCIATE FACULTY PAGE 69 4) ACADEMIC ACTIVITIES PAGE 77 5) OUTREACH PAGE 163 6) CAMPUS PAGE 173 7) STAFF PAGE 179 8) ORGANIZATION PAGE 182 9) AWARDS AND HONORS PAGE 184 Editor: Ananya Dasgupta Design: Anisha Heble Photographs: Anbujawahar iv ICTS REPORT 2013-15 | DIRECTOR’S REPORT ICTS REPORT 2013-15 | DIRECTOR’S REPORT 1 DIRECTOR’S REPORT 2 ICTS REPORT 2013-15 | DIRECTOR’S REPORT ICTS REPORT 2013-15 | DIRECTOR’S REPORT 3 Several members of our faculty have been recognized with awards, grants and fellowships within India as well as internationally. The details have been provided within the report. ICTS was awarded in 2015 the prestigious ‘Targeted Grants to Institutes’ from the Simons Foundation, USA. The AIRBUS Corporate Foundation grant for an international teaching and research chair entitled ‘Mathematics of Complex Systems’, which ICTS shares with the Centre for Applicable Mathematics, underwent a successful annual review and continues. Both these grants lend enormous encouragement to a fledgling Centre and enhance the ability of ICTS to conduct programs, invite outstanding post- docs and long term visiting professors. The new ICTS Campus has started functioning. The campus is equipped with a modern library that will provide access to books and electronic resources on advanced topics; state-of-the-art computing and networking infrastructure; a data centre with high-speed connectivity and extensive storage facility for doing big-data sciences; conference and lecture halls equipped with high-end audio-video equipment for recording and broadcasting ICTS programs; healthcare, childcare and recreational facilities for members and visitors.