Certification
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
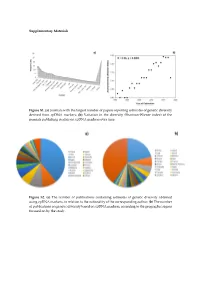
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

Appendix C Plant and Animal Species Observed
Appendix C Plant and Animal Species Observed This list includes vascular plants, mammals, birds, reptiles, and amphibians observed in the BSA by biologists during various surveys in 2005 and 2006. This list does not include invertebrate species. Invertebrates that would be most commonly encountered on the site would include butterflies, flies, dragonflies, damselflies, beetles, earwigs, grasshoppers, crickets, termites, true bugs, mantids, lacewings, bees, wasps, ants, and spiders. PLANT SPECIES OBSERVED Scientific Name Common Name Invasive Plant Rating CLUB AND SPIKE MOSSES Selaginellaceae Spike moss family Selaginella cinerascens Mesa spikemoss TRUE FERNS Azollaceae Mosquito fern family Azolla filiculoides Pacific mosquito fern Marsileaceae Marsilea family Marsilea vestita Hairy waterclover Pilularia americana American pillwort Pteridaceae Lip fern family Pellaea andromedifolia Coffee fern Pentagramma triangularis Goldenback fern PINOPHYTA GYMNOSPERMS Cupressaceace Cypress family Juniperus californica California juniper DICOT FLOWERING PLANTS Aizoaceae Carpet weed family Carpobrotus edulis* Hottentot-fig HIGH Mesembryanthemum nodiflorum* Slender-leaved ice plant Trianthema portulacastrum Horse-purslane Amaranthaceae Amaranth family Amaranthus albus* Tumbling pigweed Amaranthus palmeri Palmer’s pigweed Amaranthus sp. Pigweed Anacardiaceae Sumac family Malosma laurina Laurel sumac Rhus ovata Sugar bush Rhus trilobata Skunkbush sumac Schinus molle* Peruvian pepper tree LIMITED Schinus terebinthifolius* Brazilian pepper tree LIMITED Mid -

Shared Flora of the Alta and Baja California Pacific Islands
Monographs of the Western North American Naturalist Volume 7 8th California Islands Symposium Article 12 9-25-2014 Island specialists: shared flora of the Alta and Baja California Pacific slI ands Sarah E. Ratay University of California, Los Angeles, [email protected] Sula E. Vanderplank Botanical Research Institute of Texas, 1700 University Dr., Fort Worth, TX, [email protected] Benjamin T. Wilder University of California, Riverside, CA, [email protected] Follow this and additional works at: https://scholarsarchive.byu.edu/mwnan Recommended Citation Ratay, Sarah E.; Vanderplank, Sula E.; and Wilder, Benjamin T. (2014) "Island specialists: shared flora of the Alta and Baja California Pacific slI ands," Monographs of the Western North American Naturalist: Vol. 7 , Article 12. Available at: https://scholarsarchive.byu.edu/mwnan/vol7/iss1/12 This Monograph is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Monographs of the Western North American Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Monographs of the Western North American Naturalist 7, © 2014, pp. 161–220 ISLAND SPECIALISTS: SHARED FLORA OF THE ALTA AND BAJA CALIFORNIA PACIFIC ISLANDS Sarah E. Ratay1, Sula E. Vanderplank2, and Benjamin T. Wilder3 ABSTRACT.—The floristic connection between the mediterranean region of Baja California and the Pacific islands of Alta and Baja California provides insight into the history and origin of the California Floristic Province. We present updated species lists for all California Floristic Province islands and demonstrate the disjunct distributions of 26 taxa between the Baja California and the California Channel Islands. -

The Flowering Plants and Ferns of Anacapa Island, California
Western North American Naturalist Volume 78 Number 4 Papers from the 9th California Article 17 Islands Symposium (Part 2) 2-15-2018 The flowering plants and ernsf of Anacapa Island, California Steve Junak Santa Barbara Botanic Garden, Santa Barbara, CA, [email protected] Ralph Philbrick Follow this and additional works at: https://scholarsarchive.byu.edu/wnan Recommended Citation Junak, Steve and Philbrick, Ralph (2018) "The flowering plants and ernsf of Anacapa Island, California," Western North American Naturalist: Vol. 78 : No. 4 , Article 17. Available at: https://scholarsarchive.byu.edu/wnan/vol78/iss4/17 This Article is brought to you for free and open access by the Western North American Naturalist Publications at BYU ScholarsArchive. It has been accepted for inclusion in Western North American Naturalist by an authorized editor of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Western North American Naturalist 78(4), © 2018, pp. 652 –673 The flowering plants and ferns of Anacapa Island, California STEVE JUNAK 1,* AND RALPH PHILBRICK 2 1Santa Barbara Botanic Garden, 1212 Mission Canyon Road, Santa Barbara, CA 93105 2Deceased. 29 San Marcos Trout Club, Santa Barbara, CA 93105 ABSTRACT .—The 3 islets of Anacapa Island, with a combined area of 1.1 mi 2 (2.9 km 2), lie 13 mi (20 km) off the coast of southern California. Historically, each of Anacapa’s islets has been subjected to periodic grazing by sheep, and the eastern islet has also had a sizeable population of introduced rabbits. In spite of these past perturbations, the recovery of the island’s vegetation has been remarkable since sheep removal in 1937. -

Invasive Non-Native Plant Early Detection and Rapid Response (EDRR) Targets in Western San Diego County
Invasive non-native plant Early Detection and Rapid Response (EDRR) targets in western San Diego County Report new sightings of these plants to coordinator Jason Giessow, [email protected] or iNaturalist.org or Calflora.org Version 2019-6-3 Populations ID Scientific name Common name Growth form CDFA Habitat Status (eradicated) Sheet Aegilops triuncialis Barbed goat grass Annual grass B Grassland Active EDRR target 1 (1) Yes Ageratina adenophora Eupatory Perennial forb Q Riparian Active EDRR target 4 Yes Carrichtera annua Ward's weed Annual forb A Uplands (shrub & grass) Active EDRR target 5 Yes Centaurea solstitialis Yellow star thistle Annual forb C Grassland Active EDRR target 11 (11) Yes Centaurea stoebe Spotted knapweed Annual forb A Uplands Active EDRR target 3 (3) Yes Cytisus scoparius Scotch broom Perennial shrub C Uplands (shrub & grass) Eradicated: monitoring (1) Elymus caput-medusae Medusahead Annual grass C Grassland Active EDRR target 7 Yes Perennial sub- Enchylaena tomentosa Ruby saltbush A Uplands (shrub & grass) Assessing 1 (1) Yes shrub Euphorbia terracina Carnation spurge Annual forb B Uplands Eradicated: monitoring 8 (1) Yes Yes Euphorbia virgata Leafy spurge Annual forb A Uplands Active EDRR target 1 (1) (See ET) Genista monosperma Bridal broom Perennial shrub B Uplands (shrub & grass) Active EDRR target 4 Yes Genista monspessulana French broom Perennial shrub C Riparian or uplands Active EDRR target 5 Yes Canary Island St. Hypericum canariense Perennial shrub B Shrublands Active EDRR target 12 Yes John's wort Perennial -

Checklist of the Vascular Plants of San Diego County 5Th Edition
cHeckliSt of tHe vaScUlaR PlaNtS of SaN DieGo coUNty 5th edition Pinus torreyana subsp. torreyana Downingia concolor var. brevior Thermopsis californica var. semota Pogogyne abramsii Hulsea californica Cylindropuntia fosbergii Dudleya brevifolia Chorizanthe orcuttiana Astragalus deanei by Jon P. Rebman and Michael G. Simpson San Diego Natural History Museum and San Diego State University examples of checklist taxa: SPecieS SPecieS iNfRaSPecieS iNfRaSPecieS NaMe aUtHoR RaNk & NaMe aUtHoR Eriodictyon trichocalyx A. Heller var. lanatum (Brand) Jepson {SD 135251} [E. t. subsp. l. (Brand) Munz] Hairy yerba Santa SyNoNyM SyMBol foR NoN-NATIVE, NATURaliZeD PlaNt *Erodium cicutarium (L.) Aiton {SD 122398} red-Stem Filaree/StorkSbill HeRBaRiUM SPeciMeN coMMoN DocUMeNTATION NaMe SyMBol foR PlaNt Not liSteD iN THE JEPSON MANUAL †Rhus aromatica Aiton var. simplicifolia (Greene) Conquist {SD 118139} Single-leaF SkunkbruSH SyMBol foR StRict eNDeMic TO SaN DieGo coUNty §§Dudleya brevifolia (Moran) Moran {SD 130030} SHort-leaF dudleya [D. blochmaniae (Eastw.) Moran subsp. brevifolia Moran] 1B.1 S1.1 G2t1 ce SyMBol foR NeaR eNDeMic TO SaN DieGo coUNty §Nolina interrata Gentry {SD 79876} deHeSa nolina 1B.1 S2 G2 ce eNviRoNMeNTAL liStiNG SyMBol foR MiSiDeNtifieD PlaNt, Not occURRiNG iN coUNty (Note: this symbol used in appendix 1 only.) ?Cirsium brevistylum Cronq. indian tHiStle i checklist of the vascular plants of san Diego county 5th edition by Jon p. rebman and Michael g. simpson san Diego natural history Museum and san Diego state university publication of: san Diego natural history Museum san Diego, california ii Copyright © 2014 by Jon P. Rebman and Michael G. Simpson Fifth edition 2014. isBn 0-918969-08-5 Copyright © 2006 by Jon P. -

(12) Patent Application Publication (10) Pub. No.: US 2017/0051296A1 BEETHAM Et Al
US 20170051296A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2017/0051296A1 BEETHAM et al. (43) Pub. Date: Feb. 23, 2017 (54) METHODS AND COMPOSITIONS FOR (60) Provisional application No. 61/953,333, filed on Mar. INCREASING EFFICIENCY OF TARGETED 14, 2014, provisional application No. 62/051,579, GENE MODIFICATION USING filed on Sep. 17, 2014, provisional application No. OLGONUCLEOTDE-MEDIATED GENE 62/075,811, filed on Nov. 5, 2014, provisional appli REPAIR cation No. 62/075,816, filed on Nov. 5, 2014, provi sional application No. 62/133,129, filed on Mar. 13, (71) Applicants: CIBUS US LLC, San Diego, CA (US); 2015, provisional application No. 61/801,333, filed CIBUS EUROPE B.V., AD Kapelle on Mar. 15, 2013. (NL) (72) Inventors: Peter R. BEETHAM, Carlsbad, CA Publication Classification (US); Gregory F.W. GOCAL, San (51) Int. C. Diego, CA (US); Christian CI2N 5/82 SCHOPKE, Carlsbad, CA (US); Noel (2006.01) SAUER, Oceanside, CA (US); James (52) U.S. C. PEARCE, La Jolla, CA (US); Rosa E. CPC ....... CI2N 15/8213 (2013.01); C12N 2800/80 SEGAMI, Escondio, CA (US); Jerry (2013.01) MOZORUK, Encinitas, CA (US) (57) ABSTRACT (21) Appl. No.: 15/069.885 Provided herein include methods and compositions for (22) Filed: Mar. 14, 2016 effecting a targeted genetic change in DNA in a cell. Certain aspects and embodiments relate to improving the efficiency Related U.S. Application Data of the targeting of modifications to specific locations in (63) Continuation-in-part of application No. PCT/ genomic or other nucleotide sequences. As described herein, US2015/020622, filed on Mar. -

Thu Llultellittur
THU US009957515B2LLULTELLITTUR (12 ) United States Patent ( 10 ) Patent No. : US 9 ,957 ,515 B2 Beetham et al . ( 45) Date of Patent: May 1 , 2018 (54 ) METHODS AND COMPOSITIONS FOR 5 ,008 ,200 A 4 / 1991 Ranch et al. 5 , 024 , 944 A 6 / 1991 Collins et al . TARGETED GENE MODIFICATION 5 , 100 ,792 A 3 / 1992 Sanford et al. 5 , 204 ,253 A 4 / 1993 Sanford et al. (71 ) Applicants :CIBUS US LLC , San Diego , CA (US ) ; 5 , 219 ,746 A 6 / 1993 Brinegar et al . CIBUS EUROPE B . V ., AD Kapelle 5 , 268 ,463 A 12 / 1993 Jefferson (NL ) 5 , 302 , 523 A 4 / 1994 Coffee et al. 5 ,334 , 711 A 8 / 1994 Sproat et al. 5 , 380 , 831 A 1 / 1995 Adang et al . ( 72 ) Inventors: Peter R . Beetham , Carlsbad , CA (US ) ; 5 , 399 ,680 A 3 / 1995 Zhu et al. Gregory F . W . Gocal , San Diego , CA 5 ,424 ,412 A 6 / 1995 Brown et al . ( US ) ; Christian Schopke , Carlsbad , CA 5 ,436 , 391 A 7 / 1995 Fujimoto et al . (US ) ; Noel Sauer , Oceanside, CA (US ) ; 5 , 466 ,785 A 11/ 1995 De Framond James Pearce , La Jolla , CA (US ) ; Rosa 5 , 569, 597 A 10 / 1996 Grimsley et al . 5 ,593 , 874 A 1 / 1997 Brown et al . E . Segami, Escondio , CA (US ) ; Jerry 5 ,604 , 121 A 2 / 1997 Hilder et al . Mozoruk , Encinitas, CA (US ) 5 ,608 , 142 A 3 / 1997 Barton et al. 5 ,608 , 144 A 3 / 1997 Baden et al . ( 73 ) Assignees : CIBUS US LLC , San Diego , CA (US ) ; 5 , 608 , 149 A 3 / 1997 Barry et al. -

2018 BIODIVERSITY REPORT City of Los Angeles
2018 BIODIVERSITY REPORT City of Los Angeles Appendix B: Singapore Index Methods for Los Angeles Prepared by: Isaac Brown Ecology Studio and LA Sanitation & Environment Appendix B Singapore Index Detailed Methods Appendix B Table of Contents Appendix B Table of Contents ...................................................................................................... i Appendix B1: Singapore Index Indicator 1 ................................................................................. 1 Appendix B2: Singapore Index Indicator 2 ................................................................................. 6 Appendix B3: Singapore Index Indicator 3 ................................................................................10 Appendix B4: Singapore Index Indicator 4 ................................................................................16 Appendix B5: Singapore Index Indicator 5 ................................................................................39 Appendix B6: Singapore Index Indicator 6 ................................................................................44 Appendix B7: Singapore Index Indicator 7 ................................................................................52 Appendix B8: Singapore Index Indicator 8 ................................................................................55 Appendix B9: Singapore Index Indicator 9 ................................................................................58 Appendix B10: Singapore Index Indicator 10 ............................................................................61 -

Rare Plants of Santa Barbara County
Rare Plants of Santa Barbara County Central Coast Center for Plant Conservation Santa Barbara Botanic Garden (SBBG) 1212 Mission Canyon Rd, Santa Barbara 93105 The purpose of this list is to bring attention to those vascular plant taxa with a limited distribution in Santa Barbara County, irrespective of their status, whether they are common elsewhere or whether they are considered imperiled, threatened, or endangered by resource management agencies. This list was prepared from records maintained as part of a specimen-based database at the Santa Barbara Botanic Garden. It includes plants from the mainland and four California Channel Islands (Santa Barbara, Santa Cruz, Santa Rosa, and San Miguel). Records were primarily acquired from verified herbarium specimens deposited at several herbaria, including the University of California at Berkeley (JEPS, UC), the California Academy of Sciences (CAS, DS), the University of California at Santa Barbara (UCSB), and the Santa Barbara Botanic Garden (SBBG). Additional records were acquired from peer-reviewed publications and professional reports that refer to specimens at other herbaria or verified observations (e.g., California Natural Diversity Database). Nomenclature follows The Jepson Manual (Baldwin et al., 2012), except for addenda published on the Jepson Herbarium Online Interchange (ucjeps.berkeley.edu/jepson_flora_project.html) and recent scientific publications (e.g., Flora of North America, 1999-2007, Oxford University Press). Additional information, including nomenclature and distributional records based on documented specimens, can be accessed on the Jepson Herbarium Online Interchange. Occurrences. Any two documented locations that were estimated to be more than 1 km apart are considered to represent separate “occurrences”. Only those species, subspecies, and varieties represented by 1-8 documented natural occurrences in Santa Barbara County are listed here. -
North American Wild Relatives of Grain Crops
North Central Regional Plant Introduction NCRPIS Publications and Papers Station 2019 North American Wild Relatives of Grain Crops David M. Brenner Iowa State University, [email protected] Harold E. Bockelman U.S. Department of Agriculture Karen A. Williams U.S. Department of Agriculture Follow this and additional works at: https://lib.dr.iastate.edu/ncrpis_pubs Part of the Agricultural Science Commons, Agriculture Commons, and the Plant Breeding and Genetics Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ ncrpis_pubs/89. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Book Chapter is brought to you for free and open access by the North Central Regional Plant Introduction Station at Iowa State University Digital Repository. It has been accepted for inclusion in NCRPIS Publications and Papers by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. North American Wild Relatives of Grain Crops Abstract The wild-growing relatives of the grain crops are useful for long-term worldwide crop improvement research. There are neglected examples that should be accessioned as living seeds in gene banks. Some of the grain crops, amaranth, barnyard millet, proso millet, quinoa, and foxtail millet, have understudied unique and potentially useful crop wild relatives in North America. Other grain crops, barley, buckwheat, and oats, have fewer relatives in North America that are mostly weeds from other continents with more diverse crop wild relatives. The expanding abilities of genomic science are a reason to accession the wild species since there are improved ways to study evolution within genera and make use of wide gene pools. -

Terrestrial Vegetation Monitoring Channel Islands National Park 1996–2000 Report
NATIONAL PARK SERVICE CHANNEL ISLANDS NATIONAL PARK TECHNICAL REPORT 01-06 TERRESTRIAL VEGETATION MONITORING CHANNEL ISLANDS NATIONAL PARK 1996–2000 REPORT Lauren Johnson & Dirk Rodriguez Channel Islands National Park 1901 Spinnaker Dr. Ventura, CA 93001 Voice 805-658-5751 Fax 805-658-5798 E-mail [email protected] SEPTEMBER 2001 Terrestrial Vegetation Monitoring 1996-2000 Report ABSTRACT Channel Islands National Park began a long-term plant community monitoring program on Anacapa, Santa Barbara, and San Miguel Islands in 1984. Monitoring on Santa Rosa Island began in 1990 and on East Santa Cruz Island in 1998. Species present at each of 100 points along 163, 30-m permanent transects, are recorded annually. The number of transects on each island is determined by the island’s size and the diversity of its vegetation. Anacapa Island has 16 transects, Santa Barbara Island has 24, San Miguel Island has 16, Santa Rosa Island has 86, and East Santa Cruz Island currently has 21. This report presents the data, summarized by transect, that were collected on all five islands from 1984 - 2000. Although data from 1984 to 1995 were previously presented in Technical Report 98-08, inclusion of those data in this report presents a more complete picture for comparative purposes. For analysis of vegetation trends, the transect data should be aggregated by community types. Relative frequency of natives vs. exotics, perennials vs. annuals, and comparison of graminoids, herbs, sub-shrubs, shrubs, and trees can be derived from the data. This report supplements Technical Report 98-08, Terrestrial Vegetation Monitoring, Channel Islands National Park, 1984-1995 Report.