Poster SLC Family Cards
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Unc93b Antibody (Pab)
21.10.2014Unc93b antibody (pAb) Rabbit Anti -Human/Mouse/Rat Unc93b Instruction Manual Catalog Number PK-AB718-4553 Synonyms Unc93b Antibody: Unc93b1, homolog of C. elegans Unc93 Description The endoplasmic reticulum (ER) protein Unc93b, a human homolog of the C. elegans Unc93 gene, was initially identified by a forward genetic screen using N-ethyl-N-nitrosourea where a histidine- to-arginine substitution in Unc93b caused defects in Toll-like receptor (TLR) 3, 7 and 9 signaling. Unlike Unc93a, another homolog of the C. elegans Unc93 gene whose function is unknown, Unc93b specifically interacts with TLR3, 7 and 9; the histidine-to-arginine point mutation used to identify Unc93b abolishes this interaction. Mice carrying this point mutation are highly susceptible to infection with a number of viruses, indicating that Unc93b plays an important role in innate immunity. Multiple isoforms of Unc93a are known to exist. This antibody will not cross-react with Unc93a. Quantity 100 µg Source / Host Rabbit Immunogen Unc93b antibody was raised in rabbits against a 19 amino acid peptide from near the amino terminus of human Unc93b. Purification Method Affinity chromatography purified via peptide column. Clone / IgG Subtype Polyclonal antibody Species Reactivity Human, Mouse, Rat Specificity Multiple isoforms of Unc93a are known to exist. This antibody will not cross-react with Unc93a. Formulation Antibody is supplied in PBS containing 0.02% sodium azide. Reconstitution During shipment, small volumes of antibody will occasionally become entrapped in the seal of the product vial. For products with volumes of 200 μl or less, we recommend gently tapping the vial on a hard surface or briefly centrifuging the vial in a tabletop centrifuge to dislodge any liquid in the container’s cap. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Rhbg and Rhcg, the Putative Ammonia Transporters, Are Expressed in the Same Cells in the Distal Nephron
ARTICLES J Am Soc Nephrol 14: 545–554, 2003 RhBG and RhCG, the Putative Ammonia Transporters, Are Expressed in the Same Cells in the Distal Nephron FABIENNE QUENTIN,* DOMINIQUE ELADARI,*† LYDIE CHEVAL,‡ CLAUDE LOPEZ,§ DOMINIQUE GOOSSENS,§ YVES COLIN,§ JEAN-PIERRE CARTRON,§ MICHEL PAILLARD,*† and RE´ GINE CHAMBREY* *Institut National de la Sante´et de la Recherche Me´dicale Unite´356, Institut Fe´de´ratif de Recherche 58, Universite´Pierre et Marie Curie, Paris, France; †Hoˆpital Europe´en Georges Pompidou, Assistance Publique- Hoˆpitaux de Paris, Paris, France; ‡Centre National de la Recherche Scientifique FRE 2468, Paris, France; and §Institut National de la Sante´et de la Recherche Me´dicale Unite´76, Institut National de la Transfusion Sanguine, Paris, France. Abstract. Two nonerythroid homologs of the blood group Rh RhBG expression in distal nephron segments within the cortical proteins, RhCG and RhBG, which share homologies with specific labyrinth, medullary rays, and outer and inner medulla. RhBG ammonia transporters in primitive organisms and plants, could expression was restricted to the basolateral membrane of epithelial represent members of a new family of proteins involved in am- cells. The same localization was observed in rat and mouse monia transport in the mammalian kidney. Consistent with this kidney. RT-PCR analysis on microdissected rat nephron segments hypothesis, the expression of RhCG was recently reported at the confirmed that RhBG mRNAs were chiefly expressed in CNT apical pole of all connecting tubule (CNT) cells as well as in and cortical and outer medullary CD. Double immunostaining intercalated cells of collecting duct (CD). To assess the localiza- with RhCG demonstrated that RhBG and RhCG were coex- tion along the nephron of RhBG, polyclonal antibodies against the pressed in the same cells, but with a basolateral and apical local- Rh type B glycoprotein were generated. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -

The Role of the Renal Ammonia Transporter Rhcg in Metabolic Responses to Dietary Protein
BASIC RESEARCH www.jasn.org The Role of the Renal Ammonia Transporter Rhcg in Metabolic Responses to Dietary Protein † † † Lisa Bounoure,* Davide Ruffoni, Ralph Müller, Gisela Anna Kuhn, Soline Bourgeois,* Olivier Devuyst,* and Carsten A. Wagner* *Institute of Physiology and Zurich Center for Integrative Human Physiology, University of Zurich, Zurich, Switzerland; and †Institute for Biomechanics, ETH Zurich, Zurich, Switzerland ABSTRACT High dietary protein imposes a metabolic acid load requiring excretion and buffering by the kidney. Impaired acid excretion in CKD, with potential metabolic acidosis, may contribute to the progression of CKD. Here, we investigated the renal adaptive response of acid excretory pathways in mice to high- protein diets containing normal or low amounts of acid-producing sulfur amino acids (SAA) and examined how this adaption requires the RhCG ammonia transporter. Diets rich in SAA stimulated expression of + enzymes and transporters involved in mediating NH4 reabsorption in the thick ascending limb of the loop of Henle. The SAA-rich diet increased diuresis paralleled by downregulation of aquaporin-2 (AQP2) water + channels. The absence of Rhcg transiently reduced NH4 excretion, stimulated the ammoniagenic path- 2 way more strongly, and further enhanced diuresis by exacerbating the downregulation of the Na+/K+/2Cl cotransporter (NKCC2) and AQP2, with less phosphorylation of AQP2 at serine 256. The high protein acid load affected bone turnover, as indicated by higher Ca2+ and deoxypyridinoline excretion, phenomena exaggerated in the absence of Rhcg. In animals receiving a high-protein diet with low SAA content, the + kidney excreted alkaline urine, with low levels of NH4 and no change in bone metabolism. -

Supplemental Table 1: Genes That Show Altered Expression in Hepg2 Cells in the Presence of Exogenously Added Let-7
Supplemental Table 1: Genes that show altered expression in HepG2 cells in the presence of exogenously added let-7 Gene Title Gene Symbol RefSeq Transcriptp- IDvalue(TREAp-TMENTvalue(Let7bS) - negativLog2 Reatio control1) (Let7b - negativp-value(Let7be control1) - negativLog2 Reatio control2) (Let7b - negative control2) aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) AKR1D1 NM_005989 3.28E-12 2.52E-12 -3.85007 3.59E-12 -3.73727 lin-28 homolog B (C. elegans) LIN28B NM_001004317 6.13E-15 8.29E-15 -3.29879 1.55E-15 -3.79656 high mobility group AT-hook 2 /// high mobility group AT-hook 2 HMGA2 NM_001015886 /// NM_0034833.74E-14 /// NM_0034844.29E-14 -3.06085 4.56E-14 -3.04538 HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 HECW2 NM_020760 1.27E-13 6.65E-13 -2.94724 4.47E-12 -2.50907 cell division cycle 25A CDC25A NM_001789 /// NM_2015672.01E-11 7.32E-11 -2.88831 1.99E-11 -3.22735 hypothetical protein FLJ21986 FLJ21986 NM_024913 1.05E-09 5.19E-10 -2.80277 1.18E-09 -2.61084 solute carrier family 2 (facilitated glucose transporter), member 3 SLC2A3 NM_006931 1.59E-13 3.49E-13 -2.78111 1.84E-12 -2.41734 Transcribed locus --- --- 2.58E-13 1.08E-13 -2.59794 1.69E-13 -2.50248 Hypothetical protein LOC145786 LOC145786 --- 4.23E-12 1.07E-11 -2.58849 3.00E-12 -2.88135 Dicer1, Dcr-1 homolog (Drosophila) DICER1 NM_030621 /// NM_1774381.06E-08 4.37E-09 -2.5442 4.49E-09 -2.53796 mannose-binding lectin (protein C) 2, soluble (opsonic defect) MBL2 NM_000242 9.73E-10 1.48E-09 -2.53211 9.84E-10 -2.62363 cell -

Cloning and Functional Characterization of Putative Heavy Metal Stress Responsive (Echmr) Gene from Eichhornia Crassipes (Solm L.)
Cloning and Functional Characterization of Putative Heavy Metal Stress responsive (Echmr) gene from Eichhornia crassipes (Solm L.) A thesis submitted to INDIAN INSTITUTE OF TECHNOLOGY GUWAHATI For the award of the degree of DOCTOR OF PHILOSOPHY IN BIOTECHNOLOGY By GANESH THAPA Department of Biotechnology Indian Institute of Technology Guwahati December 2012 INDIAN INSTITUTE OF TECHNOLOGY GUWAHATI DEPARTMENT OF BIOTECHNOLOGY STATEMENT I hereby declare that the work presented in this thesis is original and was obtained from the studies undertaken by me in the Department of Biotechnology, Indian Institute of Technology Guwahati, India, under the supervision of Prof. Lingaraj Sahoo. As per the general norms of reporting research findings, due acknowledgements have been made wherever the research findings of other researchers have been cited in this thesis. Ganesh Thapa Indian Institute of Technology Guwahati, Guwahati December 31st, 2012 TH-1156_08610609 DEPARTMENT OF BIOTECHNOLOGY INDIAN INSTITUTE OF TECHNOLOGY GUWAHATI GUWAHATI-781 039 _____________________________________________________________________ CERTIFICATE This is to certify that the work presented in the form of a thesis in fulfillment of the requirement for the award of the Ph.D degree of The Indian Institute of Technology Guwahati by Ganesh Thapa is his original work. The matter presented in this thesis incorporates the findings of independent research work carried out by the researcher himself. The entire research work and the thesis have been built up under my supervision. The matter contained in this thesis has not been submitted elsewhere for the award of any other degree. CERTIFIED Date: 31st Dec. 2012 Prof. Lingaraj Sahoo (Thesis Supervisor) Department of Biotechnology Indian Institute of Technology Guwahati Guwahati- 781 039 Assam, INDIA TH-1156_08610609 .………. -

Fabbri Et Al. Whole Genome Analysis and Micrornas Regulation in Hepg2 Cells Exposed to Cadmium Supplementary Data
Fabbri et al. Whole Genome Analysis and MicroRNAs Regulation in HepG2 Cells Exposed to Cadmium Supplementary Data Tab. S1: KEGG enrichment for downregulated genes Genes identified in Figure 1 were analyzed by DAVID for associations with particular KEGG pathways. KEGG Entry is KEGG identifier, Name is name of the KEGG pathway, Genes shows the number of genes associated with the specific pathway, the PValue refers to how significant an association a particular KEGG pathway has with the gene list. KEGG Entry Name Genes PValue hsa04610 Complement and coagulation cascades 22 1.11E-14 hsa00260 Glycine, serine and threonine metabolism 11 8.50E-08 hsa00071 Fatty acid metabolism 11 9.41E-07 hsa00650 Butanoate metabolism 9 1.89E-05 hsa00100 Steroid biosynthesis 7 2.09E-05 hsa00280 Valine, leucine and isoleucine degradation 10 2.47E-05 hsa00380 Tryptophan metabolism 9 8.40E-05 hsa00330 Arginine and proline metabolism 10 1.16E-04 hsa00900 Terpenoid backbone biosynthesis 6 1.46E-04 hsa00980 Metabolism of xenobiotics by cytochrome P450 10 2.71E-04 hsa00010 Glycolysis / Gluconeogenesis 10 2.71E-04 hsa00982 Drug metabolism 10 3.98E-04 hsa03320 PPAR signaling pathway 10 7.98E-04 hsa00620 Pyruvate metabolism 7 0.003185725 hsa00561 Glycerolipid metabolism 7 0.005184764 hsa00640 Propanoate metabolism 6 0.005876295 hsa00910 Nitrogen metabolism 5 0.009266837 hsa00480 Glutathione metabolism 7 0.009722623 hsa04950 Maturity onset diabetes of the young 5 0.012498995 hsa00903 Limonene and pinene degradation 4 0.013441968 hsa00680 Methane metabolism 3 0.018538005 hsa00120 Primary bile acid biosynthesis 4 0.01958794 hsa00340 Histidine metabolism 5 0.020928876 hsa00310 Lysine degradation 6 0.022199526 hsa00250 Alanine, aspartate and glutamate metabolism 5 0.026189764 hsa00410 beta-Alanine metabolism 4 0.04583419 hsa01040 Biosynthesis of unsaturated fatty acids 4 0.04583419 ALTEX, 2/12 SUPPL., 1 FABBRI ET AL . -

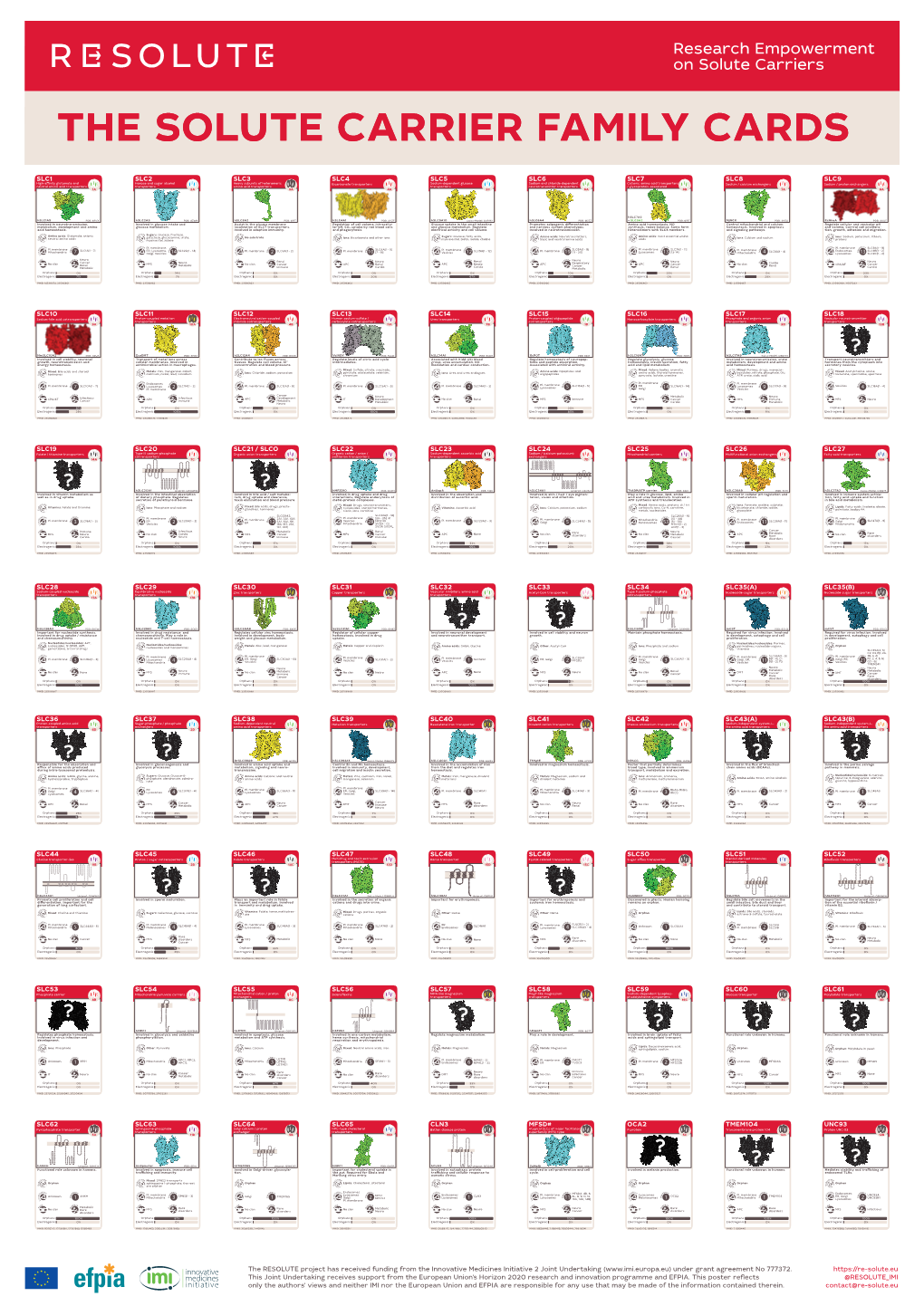
The Genetic Landscape of the Human Solute Carrier (SLC) Transporter Superfamily
Human Genetics (2019) 138:1359–1377 https://doi.org/10.1007/s00439-019-02081-x ORIGINAL INVESTIGATION The genetic landscape of the human solute carrier (SLC) transporter superfamily Lena Schaller1 · Volker M. Lauschke1 Received: 4 August 2019 / Accepted: 26 October 2019 / Published online: 2 November 2019 © The Author(s) 2019 Abstract The human solute carrier (SLC) superfamily of transporters is comprised of over 400 membrane-bound proteins, and plays essential roles in a multitude of physiological and pharmacological processes. In addition, perturbation of SLC transporter function underlies numerous human diseases, which renders SLC transporters attractive drug targets. Common genetic polymorphisms in SLC genes have been associated with inter-individual diferences in drug efcacy and toxicity. However, despite their tremendous clinical relevance, epidemiological data of these variants are mostly derived from heterogeneous cohorts of small sample size and the genetic SLC landscape beyond these common variants has not been comprehensively assessed. In this study, we analyzed Next-Generation Sequencing data from 141,456 individuals from seven major human populations to evaluate genetic variability, its functional consequences, and ethnogeographic patterns across the entire SLC superfamily of transporters. Importantly, of the 204,287 exonic single-nucleotide variants (SNVs) which we identifed, 99.8% were present in less than 1% of analyzed alleles. Comprehensive computational analyses using 13 partially orthogonal algorithms that predict the functional impact of genetic variations based on sequence information, evolutionary conserva- tion, structural considerations, and functional genomics data revealed that each individual genome harbors 29.7 variants with putative functional efects, of which rare variants account for 18%. Inter-ethnic variability was found to be extensive, and 83% of deleterious SLC variants were only identifed in a single population. -

Supplementary Methods
Supplementary methods Human lung tissues and tissue microarray (TMA) All human tissues were obtained from the Lung Cancer Specialized Program of Research Excellence (SPORE) Tissue Bank at the M.D. Anderson Cancer Center (Houston, TX). A collection of 26 lung adenocarcinomas and 24 non-tumoral paired tissues were snap-frozen and preserved in liquid nitrogen for total RNA extraction. For each tissue sample, the percentage of malignant tissue was calculated and the cellular composition of specimens was determined by histological examination (I.I.W.) following Hematoxylin-Eosin (H&E) staining. All malignant samples retained contained more than 50% tumor cells. Specimens resected from NSCLC stages I-IV patients who had no prior chemotherapy or radiotherapy were used for TMA analysis by immunohistochemistry. Patients who had smoked at least 100 cigarettes in their lifetime were defined as smokers. Samples were fixed in formalin, embedded in paraffin, stained with H&E, and reviewed by an experienced pathologist (I.I.W.). The 413 tissue specimens collected from 283 patients included 62 normal bronchial epithelia, 61 bronchial hyperplasias (Hyp), 15 squamous metaplasias (SqM), 9 squamous dysplasias (Dys), 26 carcinomas in situ (CIS), as well as 98 squamous cell carcinomas (SCC) and 141 adenocarcinomas. Normal bronchial epithelia, hyperplasia, squamous metaplasia, dysplasia, CIS, and SCC were considered to represent different steps in the development of SCCs. All tumors and lesions were classified according to the World Health Organization (WHO) 2004 criteria. The TMAs were prepared with a manual tissue arrayer (Advanced Tissue Arrayer ATA100, Chemicon International, Temecula, CA) using 1-mm-diameter cores in triplicate for tumors and 1.5 to 2-mm cores for normal epithelial and premalignant lesions. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing