Halichoeres Gurrobyi, a New Labrid Fish (Teleostei: Labridae) from Mauritius in the Southwestern Indian Ocean, with a Review of the H
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reef Fishes of the Bird's Head Peninsula, West
Check List 5(3): 587–628, 2009. ISSN: 1809-127X LISTS OF SPECIES Reef fishes of the Bird’s Head Peninsula, West Papua, Indonesia Gerald R. Allen 1 Mark V. Erdmann 2 1 Department of Aquatic Zoology, Western Australian Museum. Locked Bag 49, Welshpool DC, Perth, Western Australia 6986. E-mail: [email protected] 2 Conservation International Indonesia Marine Program. Jl. Dr. Muwardi No. 17, Renon, Denpasar 80235 Indonesia. Abstract A checklist of shallow (to 60 m depth) reef fishes is provided for the Bird’s Head Peninsula region of West Papua, Indonesia. The area, which occupies the extreme western end of New Guinea, contains the world’s most diverse assemblage of coral reef fishes. The current checklist, which includes both historical records and recent survey results, includes 1,511 species in 451 genera and 111 families. Respective species totals for the three main coral reef areas – Raja Ampat Islands, Fakfak-Kaimana coast, and Cenderawasih Bay – are 1320, 995, and 877. In addition to its extraordinary species diversity, the region exhibits a remarkable level of endemism considering its relatively small area. A total of 26 species in 14 families are currently considered to be confined to the region. Introduction and finally a complex geologic past highlighted The region consisting of eastern Indonesia, East by shifting island arcs, oceanic plate collisions, Timor, Sabah, Philippines, Papua New Guinea, and widely fluctuating sea levels (Polhemus and the Solomon Islands is the global centre of 2007). reef fish diversity (Allen 2008). Approximately 2,460 species or 60 percent of the entire reef fish The Bird’s Head Peninsula and surrounding fauna of the Indo-West Pacific inhabits this waters has attracted the attention of naturalists and region, which is commonly referred to as the scientists ever since it was first visited by Coral Triangle (CT). -

Ecomorphology of Locomotion in Labrid Fishes
Environmental Biology of Fishes 65: 47–62, 2002. © 2002 Kluwer Academic Publishers. Printed in the Netherlands. Ecomorphology of locomotion in labrid fishes Peter C. Wainwrighta, David R. Bellwoodb & Mark W. Westneatc aSection of Evolution and Ecology, University of California, One Shields Avenue, Davis, CA 95616, U.S.A. (e-mail: [email protected]) bCentre for Coral Reef Biodiversity, Department of Marine Biology, James Cook University, Townsville, Queensland, Australia 4811 cDepartment of Zoology, Field Museum of Natural History, 1400 South Lakeshore Drive, Chicago, IL 60505, U.S.A. Received 16 May 2001 Accepted 17 January 2002 Key words: pectoral fin, aspect ratio, Labridae, allometry, convergent evolution, swimming speed Synopsis The Labridae is an ecologically diverse group of mostly reef associated marine fishes that swim primarily by oscillating their pectoral fins. To generate locomotor thrust, labrids employ the paired pectoral fins in motions that range from a fore-aft rowing stroke to a dorso-ventral flapping stroke. Species that emphasize one or the other behavior are expected to benefit from alternative fin shapes that maximize performance of their primary swimming behavior. We document the diversity of pectoral fin shape in 143 species of labrids from the Great Barrier Reef and the Caribbean. Pectoral fin aspect ratio ranged among species from 1.12 to 4.48 and showed a distribution with two peaks at about 2.0 and 3.0. Higher aspect ratio fins typically had a relatively long leading edge and were narrower distally. Body mass only explained 3% of the variation in fin aspect ratio in spite of four orders of magnitude range and an expectation that the advantages of high aspect ratio fins and flapping motion are greatest at large body sizes. -

Halichoeres Bivittatus (Bloch, 1791) Frequent Synonyms / Misidentifications: None / Halichoeres Maculipinna (Müller and Troschel, 1848)
click for previous page 1710 Bony Fishes Halichoeres bivittatus (Bloch, 1791) Frequent synonyms / misidentifications: None / Halichoeres maculipinna (Müller and Troschel, 1848). FAO names: En - Slippery dick. Diagnostic characters: Body slender, depth 3.3 to 4.6 in standard length.Head rounded and scaleless;snout blunt; 1 pair of enlarged canine teeth at front of upper jaw and a small canine posteriorly near corner of mouth; 2 pairs of enlarged canine teeth anteriorly in lower jaw. Gill rakers on first arch 16 to 19. Dorsal fin continu- ous, with 9 spines and 11 soft rays;anal fin with 3 spines and 9 soft rays;caudal fin rounded;pectoral-fin rays 13. Lateral line continuous with an abrupt downward bend beneath soft portion of dorsal fin, and 27 pored scales. Colour: body colour variable, primarily pale green to white ground colour with a dark midbody stripe, a second lower stripe often present but less distinct; small green and yellow bicoloured spot above pectoral fin; pinkish or orange markings on the head, these sometimes outlined with pale blue; in adults, the tips of the cau- dal-fin lobes are black. Size: Maximum length to about 20 cm. Habitat, biology, and fisheries: Inhabits a di- versity of habitats from coral reef to rocky reef and seagrass beds. Any disturbance of the bot- tom, such as the overturning of a rock will attract a swarm of them, all hoping to find food uncov- ered. Feeds omnivorously on crabs, fishes, sea urchins, polychaetes, molluscs, and brittle stars. This species is not marketed for food, but is com- monly seen in the aquarium trade. -

Universidade Tiradentes Programa De Pós - Graduação Em Saúde E Ambiente
UNIVERSIDADE TIRADENTES PROGRAMA DE PÓS - GRADUAÇÃO EM SAÚDE E AMBIENTE ASPECTOS PARASITOLÓGICOS DE RAIAS DO GÊNERO Hypanus (MYLIOBATIFORMES: DASYATIDAE) NO NORDESTE DO BRASIL MARINA GOMES LEONARDO Aracaju Maio/2018 UNIVERSIDADE TIRADENTES PROGRAMA DE PÓS - GRADUAÇÃO EM SAÚDE E AMBIENTE ASPECTOS PARASITOLÓGICOS DE RAIAS DO GÊNERO Hypanus (MYLIOBATIFORMES: DASYATIDAE) NO NORDESTE DO BRASIL Dissertação de Mestrado submetida à banca examinadora para a obtenção do título de Mestre em Saúde e Ambiente, na área de concentração Saúde e Ambiente. MARINA GOMES LEONARDO Orientadores Prof. Dr. Ricardo Massato Takemoto Prof. Dr. Rubens Riscala Madi Aracaju/SE 2018 Leonardo, Marina Gomes L581a Aspectos parasitológicos de raias do gênero hypanus (Myliobatiformes: Dasyatidae) no nordeste do Brasil / Marina Gomes Leonardo ; orientação [de] Prof. Dr. Ricardo Massato Takemoto, Prof. Dr. Rubens Riscala Madi– Aracaju: UNIT, 2018. 51f. : 30 cm Dissertação (Mestrado em Saúde e Ambiente) - Universidade Tiradentes, 2018 Inclui bibliografia. 1. Fauna parasitária. 2.Raia de Pedra. 3. Raia Lixa. 4. Zoonose. 5. Sergipe I. Leonardo, Marina Gomes. II. Takemoto, Ricardo Massato (orient.). III. Madi, Rubens Riscala (oriente.) IV. Universidade Tiradentes. V. Título. CDU: 591. 69: 597. 317.7 ASPECTOS PARASITOLÓGICOS DE RAIAS DO GÊNERO Hypanus (MYLIOBATIFORMES: DASYATIDAE) NO NORDESTE DO BRASIL Marina Gomes Leonardo Dissertação de mestrado apresentada à banca examinadora para do título de Mestre em Saúde e Ambiente, na de concentração Saúde e Ambiente. Aprovada por: Dr. Ricardo Massato Takemoto Dr. Rubens Riscala Madi Dra. Verônica de Lourdes Sierpe Geraldo Universidade Tiradentes Dra. Maria Lúcia Góes de Araújo Universidade Federal de Sergipe “Deus transforma choro em sorriso, dor em força, fraqueza em fé e SONHO em realidade.” Autor desconhecido Agradecimentos Sendo clichê e realista, gostaria de agradecer primeiramente a Deus, pois sem ele e minha fé, provavelmente não teria superado nem metade das adversidades por quais passei nesses dois anos e 3 meses. -

Chinese Red Swimming Crab (Portunus Haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018)
Chinese Red Swimming Crab (Portunus haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018) Prepared by Min Liu & Bai-an Lin Fish Biology Laboratory College of Ocean and Earth Sciences, Xiamen University March 2019 Contents 1. Introduction........................................................................................................ 5 2. Materials and Methods ...................................................................................... 6 2.1. Study site and survey frequency .................................................................... 6 2.2. Sample collection .......................................................................................... 7 2.3. Species identification................................................................................... 10 2.4. Sample measurement ................................................................................... 11 2.5. Interviews.................................................................................................... 13 2.6. Estimation of annual capture volume of Portunus haanii ............................. 15 3. Results .............................................................................................................. 15 3.1. Species diversity.......................................................................................... 15 3.1.1. Species composition .............................................................................. 15 3.1.2. ETP species ......................................................................................... -
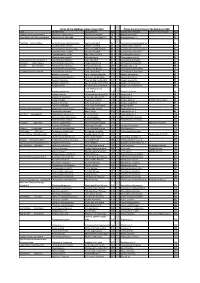
Download the Full List of Species Name Changes For
Fishes of the Maldives Indian Ocean 2014 Photo Guide to Fishes of the Maldives 1998 Family Scientific Name Common Name Page Scientific Name Changed Common Name Changed Page Solderfishes Holocentridae-2 Myripristis melanosticta Splendid Soldierfish 78 Myripristis botche 47 Ghost Pipefishes Solenostomidae Solenostomus halimeda Coralline Ghost Pipefish 81 Solenostome sp. 1 50 Pipefishes Syngnathidae Corythoichthys haematopterus Reef-top Pipefish 82 Corythoichtys haematopterus* 51 Corythoichthys schultzi Schultz’s Pipefish 83 Corythoichtys schultzi* 51 Corythoichthys flavofasciatus Yellow-banded Pipefish 83 Corythoichtys flavofasciatus* 51 Corythoichthys insularis Cheeked Pipefish 83 Corythoichtys insularis* 51 Doryrhamphus excisus Blue-stripe Pipefish 84 Doryrhamphus exicus* 53 Hippocampus jayakari Spiny Seahorse 85 Hippocampus hystrix 53 Scorpionfishes Scorpaenidae-2 Scorpaenopsis diabolus False Stonefish 89 Scorpaenopsis diabola 57 Groupers Serranidae-2 Cephalopholis leopardus Leopard Rock Cod 98 Cephalopholis leoparda 66 Basslets Serranidae-3 Pseudanthias lunulatus Yellow-eye Basslet 107 Pseudanthias n. sp. 74 Pseudanthias bimarginatus Short-snout Basslet 109 Pseudanthias parvirostris 76 Cardinalfishes Apogonidae Cheilodipterus lineatus Tiger Cardinalfish 114 Cheilodipterus macrodon 81 Apogon urostigma Spiny-head Cardinalfish 117 Apogon kallopterus 83 Apogon melanorhynchus Spiny-eye Cardinalfish 117 Apogon fraenatus 84 Apogon fraenatus Tapered-line Cardinalfish 117 Apogon exostigma 84 Apogon savayensis Big-eye Dusky-cardinalfish 118 Apogon -

An Investigation Into Australian Freshwater Zooplankton with Particular Reference to Ceriodaphnia Species (Cladocera: Daphniidae)
An investigation into Australian freshwater zooplankton with particular reference to Ceriodaphnia species (Cladocera: Daphniidae) Pranay Sharma School of Earth and Environmental Sciences July 2014 Supervisors Dr Frederick Recknagel Dr John Jennings Dr Russell Shiel Dr Scott Mills Table of Contents Abstract ...................................................................................................................................... 3 Declaration ................................................................................................................................. 5 Acknowledgements .................................................................................................................... 6 Chapter 1: General Introduction .......................................................................................... 10 Molecular Taxonomy ..................................................................................................... 12 Cytochrome C Oxidase subunit I ................................................................................... 16 Traditional taxonomy and cataloguing biodiversity ....................................................... 20 Integrated taxonomy ....................................................................................................... 21 Taxonomic status of zooplankton in Australia ............................................................... 22 Thesis Aims/objectives .................................................................................................. -

Fishery Conservation and Management Pt. 622, App. A
Fishery Conservation and Management Pt. 622, App. A vessel's unsorted catch of Gulf reef to complete prohibition), and seasonal fish: or area closures. (1) The requirement for a valid com- (g) South Atlantic golden crab. MSY, mercial vessel permit for Gulf reef fish ABC, TAC, quotas (including quotas in order to sell Gulf reef fish. equal to zero), trip limits, minimum (2) Minimum size limits for Gulf reef sizes, gear regulations and restrictions, fish. permit requirements, seasonal or area (3) Bag limits for Gulf reef fish. closures, time frame for recovery of (4) The prohibition on sale of Gulf golden crab if overfished, fishing year reef fish after a quota closure. (adjustment not to exceed 2 months), (b) Other provisions of this part not- observer requirements, and authority withstanding, a dealer in a Gulf state for the RD to close the fishery when a is exempt from the requirement for a quota is reached or is projected to be dealer permit for Gulf reef fish to re- reached. ceive Gulf reef fish harvested from the (h) South Atlantic shrimp. Certified Gulf EEZ by a vessel in the Gulf BRDs and BRD specifications. groundfish trawl fishery. [61 FR 34934, July 3, 1996, as amended at 61 FR 43960, Aug. 27, 1996; 62 FR 13988, Mar. 25, § 622.48 Adjustment of management 1997; 62 FR 18539, Apr. 16, 1997] measures. In accordance with the framework APPENDIX A TO PART 622ÐSPECIES procedures of the applicable FMPs, the TABLES RD may establish or modify the follow- TABLE 1 OF APPENDIX A TO PART 622Ð ing management measures: CARIBBEAN CORAL REEF RESOURCES (a) Caribbean coral reef resources. -

Phylogenetics and Geography of Speciation in New World Halichoeres T Wrasses ⁎ Peter C
Molecular Phylogenetics and Evolution 121 (2018) 35–45 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogenetics and geography of speciation in New World Halichoeres T wrasses ⁎ Peter C. Wainwrighta, , Francesco Santinib, David R. Bellwoodc, D. Ross Robertsond, Luiz A. Rochae, Michael E. Alfarob a Department of Evolution and Ecology, University of California, Davis, One Shields Avenue, Davis, CA 95616, USA b University of California Los Angeles, Department of Ecology and Evolutionary Biology, 610 Charles E Young Drive South, Los Angeles, CA 90095, USA c Centre of Excellence for Coral Reef Studies, James Cook University, Townsville, QLD, 4811, Australia d Smithsonian Tropical Research Institute, Balboa, Rep. de Panama, Unit 0948, APO AA 34002, USA e California Academy of Sciences, Section of Ichthyology, 55 Music Concourse Drive, San Francisco, CA 94118, USA ARTICLE INFO ABSTRACT Keywords: The New World Halichoeres comprises about 30 small to medium sized wrasse species that are prominent Labridae members of reef communities throughout the tropical Western Atlantic and Eastern Pacific. We conducted a New World Halichoeres phylogenetic analysis of this group and related lineages using new and previously published sequence data. We Western Atlantic estimated divergence times, evaluated the monophyly of this group, their relationship to other labrids, as well as Eastern Pacific the time-course and geography of speciation. These analyses show that all members of New World Halichoeres Phylogeny form a monophyletic group that includes Oxyjulis and Sagittalarva. New World Halichoeres is one of numerous Biogeography Speciation labrid groups that appear to have radiated rapidly about 32 Ma and form a large polytomy within the julidine wrasses. -

Chec List an Update to the List of Coral Reef Fishes from Koh Tao, Gulf Of
Check List 10(5): 1123–1133, 2014 © 2014 Check List and Authors Chec List ISSN 1809-127X (available at www.checklist.org.br) Journal of species lists and distribution An update to the list PECIES S Gulf of Thailand OF of coral reef fishes from Koh Tao, 1* 2 ISTS Patrick Scaps and Chad Scott L 1 Laboratoire de Biologie animale,[email protected] Université des Sciences et Technologies de Lille, 59 655 Villeneuve d’Ascq Cédex, France. 2 New Heaven Reef Conservation Program, 48 Moo 3, Koh Tao, Suratthani, Thailand, 84360. * Corresponding author: E-mail: ABSTRACT: (i.e., cryptic species or transient Twenty-one species are reported for the first time from Koh Tao (Turtle Island) in the Gulf of Thailand. TInformationhis and photographs were obtained from local scuba divers in order to censusAntennatus rare and Histrio), Ophichthyidae (genusspecies onlyCallechelys present), duringPlatycephalidae one season) (genus or not previouslyThysanophrys recorded), Plotosidae fish species (genus living Plotosus on or near) and coral Synanceiidae reefs from the(genera area. Inimicus is the and first Synanceia time that species belonging to the families AntennariidaePseudobalistes, (genera Balistidae; Cyclichthys, Diodontidae; Bolbometopon, Scaridae; and Hippocampus, ), and reef-fish genera of severalAntennatus families nummifer ( (Antennariidae), Pseudobalistes marginatus (Balistidae), Monacanthus chinensis (Monacanthidae),Syngnathidae), Callechelys among others,marmora haveta been(Ophichthyidae), recorded in KohThysnophrys Tao. Of the cf. 21 chiltonae species reported(Platycephalidae), for the first Bolbometopon time from muricatumKoh Tao, 7 (Scaridae) species ( and Synanceia cf. verrucosa (Synanceidae)) are new records for the species found in the present study. Gulf of Thailand. To date, 223 species of coral reef fishes belonging to 53 families are known from Koh Tao, including the 10.15560/10.5.1123 DOI: Introduction MaterialS and Methods 2 archipelago in the western Gulf of Thailand. -

Hermaphroditism in Fish
Tesis doctoral Evolutionary transitions, environmental correlates and life-history traits associated with the distribution of the different forms of hermaphroditism in fish Susanna Pla Quirante Tesi presentada per a optar al títol de Doctor per la Universitat Autònoma de Barcelona, programa de doctorat en Aqüicultura, del Departament de Biologia Animal, de Biologia Vegetal i Ecologia. Director: Tutor: Dr. Francesc Piferrer Circuns Dr. Lluís Tort Bardolet Departament de Recursos Marins Renovables Departament de Biologia Cel·lular, Institut de Ciències del Mar Fisiologia i Immunologia Consell Superior d’Investigacions Científiques Universitat Autònoma de Barcelona La doctoranda: Susanna Pla Quirante Barcelona, Setembre de 2019 To my mother Agraïments / Acknowledgements / Agradecimientos Vull agrair a totes aquelles persones que han aportat els seus coneixements i dedicació a fer possible aquesta tesi, tant a nivell professional com personal. Per començar, vull agrair al meu director de tesi, el Dr. Francesc Piferrer, per haver-me donat aquesta oportunitat i per haver confiat en mi des del principi. Sempre admiraré i recordaré el teu entusiasme en la ciència i de la contínua formació rebuda, tant a nivell científic com personal. Des del primer dia, a través dels teus consells i coneixements, he experimentat un continu aprenentatge que sens dubte ha derivat a una gran evolució personal. Principalment he après a identificar les meves capacitats i les meves limitacions, i a ser resolutiva davant de qualsevol adversitat. Per tant, el meu més sincer agraïment, que mai oblidaré. During the thesis, I was able to meet incredible people from the scientific world. During my stay at the University of Manchester, where I learned the techniques of phylogenetic analysis, I had one of the best professional experiences with Dr. -

NBSREA Design Cvrs V2.Pub
February 2009 TNC Pacific Island Countries Report No 1/09 Rapid Ecological Assessment Northern Bismarck Sea Papua New Guinea Technical report of survey conducted August 13 to September 7, 2006 Edited by: Richard Hamilton, Alison Green and Jeanine Almany Supported by: AP Anonymous February 2009 TNC Pacific Island Countries Report No 1/09 Rapid Ecological Assessment Northern Bismarck Sea Papua New Guinea Technical report of survey conducted August 13 to September 7, 2006 Edited by: Richard Hamilton, Alison Green and Jeanine Almany Published by: The Nature Conservancy, Indo-Pacific Resource Centre Author Contact Details: Dr. Richard Hamilton, 51 Edmondstone Street, South Brisbane, QLD 4101 Australia Email: [email protected] Suggested Citation: Hamilton, R., A. Green and J. Almany (eds.) 2009. Rapid Ecological Assessment: Northern Bismarck Sea, Papua New Guinea. Technical report of survey conducted August 13 to September 7, 2006. TNC Pacific Island Countries Report No. 1/09. © 2009, The Nature Conservancy All Rights Reserved. Reproduction for any purpose is prohibited without prior permission. Cover Photo: Manus © Gerald Allen ISBN 9980-9964-9-8 Available from: Indo-Pacific Resource Centre The Nature Conservancy 51 Edmondstone Street South Brisbane, QLD 4101 Australia Or via the worldwide web at: conserveonline.org/workspaces/pacific.island.countries.publications ii Foreword Manus and New Ireland provinces lie north of the Papua New Guinea mainland in the Bismarck Archipelago. More than half of the local communities in our provinces are coastal inhabitants, who for thousands of years have depended on marine resources for their livelihood. For coastal communities survival and prosperity is integrally linked to healthy marine ecosystems.