Afrostyrax Lepidophyllus Extracts Exhibit in Vitro Free Radical
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alphabetical Lists of the Vascular Plant Families with Their Phylogenetic
Colligo 2 (1) : 3-10 BOTANIQUE Alphabetical lists of the vascular plant families with their phylogenetic classification numbers Listes alphabétiques des familles de plantes vasculaires avec leurs numéros de classement phylogénétique FRÉDÉRIC DANET* *Mairie de Lyon, Espaces verts, Jardin botanique, Herbier, 69205 Lyon cedex 01, France - [email protected] Citation : Danet F., 2019. Alphabetical lists of the vascular plant families with their phylogenetic classification numbers. Colligo, 2(1) : 3- 10. https://perma.cc/2WFD-A2A7 KEY-WORDS Angiosperms family arrangement Summary: This paper provides, for herbarium cura- Gymnosperms Classification tors, the alphabetical lists of the recognized families Pteridophytes APG system in pteridophytes, gymnosperms and angiosperms Ferns PPG system with their phylogenetic classification numbers. Lycophytes phylogeny Herbarium MOTS-CLÉS Angiospermes rangement des familles Résumé : Cet article produit, pour les conservateurs Gymnospermes Classification d’herbier, les listes alphabétiques des familles recon- Ptéridophytes système APG nues pour les ptéridophytes, les gymnospermes et Fougères système PPG les angiospermes avec leurs numéros de classement Lycophytes phylogénie phylogénétique. Herbier Introduction These alphabetical lists have been established for the systems of A.-L de Jussieu, A.-P. de Can- The organization of herbarium collections con- dolle, Bentham & Hooker, etc. that are still used sists in arranging the specimens logically to in the management of historical herbaria find and reclassify them easily in the appro- whose original classification is voluntarily pre- priate storage units. In the vascular plant col- served. lections, commonly used methods are systema- Recent classification systems based on molecu- tic classification, alphabetical classification, or lar phylogenies have developed, and herbaria combinations of both. -

Tree Composition and Ecological Structure of Akak Forest Area
Environment and Natural Resources Research; Vol. 9, No. 4; 2019 ISSN 1927-0488 E-ISSN 1927-0496 Published by Canadian Center of Science and Education Tree Composition and Ecological Structure of Akak Forest Area Agbor James Ayamba1,2, Nkwatoh Athanasius Fuashi1, & Ayuk Elizabeth Orock1 1 Department of Environmental Science, University of Buea, Cameroon 2 Ajemalebu Self Help, Kumba, South West Region, Cameroon Correspondence: Agbor James Ayamba, Department of Environmental Science, University of Buea, Cameroon. Tel: 237-652-079-481. E-mail: [email protected] Received: August 2, 2019 Accepted: September 11, 2019 Online Published: October 12, 2019 doi:10.5539/enrr.v9n4p23 URL: https://doi.org/10.5539/enrr.v9n4p23 Abstract Tree composition and ecological structure were assessed in Akak forest area with the objective of assessing the floristic composition and the regeneration potentials. The study was carried out between April 2018 to February 2019. A total of 49 logged stumps were selected within the Akak forest spanning a period of 5 years and 20m x 20m transects were demarcated. All plants species <1cm and above were identified and recorded. Results revealed that a total of 5239 individuals from 71 families, 216 genera and 384species were identified in the study area. The maximum plants species was recorded in the year 2015 (376 species). The maximum number of species and regeneration potentials was found in the family Fabaceae, (99 species) and (31) respectively. Baphia nitida, Musanga cecropioides and Angylocalyx pynaertii were the most dominant plants specie in the years 2013, 2015 and 2017 respectively. The year 2017 depicts the highest Simpson diversity with value of (0.989) while the year 2015 show the highest Simpson dominance with value of (0.013). -

Research Article
Research Article: Maximum Likelihood Analyses of 3,490 rbcL Sequences: Scalability of Comprehensive Inference versus Group-Specific Taxon Sampling Alexandros Stamatakis1,*, Markus Göker2,3, Guido Grimm4 1 The Exelixis Lab, Dept. of Computer Science, Technische Universität München, Germany 2 Organismic Botany, Eberhard-Karls-University, Tübingen, Germany 3 German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany 4 Department of Palaeobotany, Swedish Museum of Natural History, Stockholm, Sweden * Corresponding author: Technische Universität München, Fakultät für Informatik, I 12, Boltzmannstr. 3, 85748 Garching b. München, Tel: +49 89 28919434, Fax: +49 89 28919414, Email: [email protected] Keywords: Phylogenetic Inference, Maximum Likelihood, RAxML, large single–gene datasets, eudicots Running head: Scalability of Maximum Likelihood Analyses Abbreviations: GRTS: Group-based Randomized Taxon-Subsampling; TU: Taxonomic Unit. Abstract The constant accumulation of sequence data poses new computational and methodological challenges for phylogenetic inference, since multiple sequence alignments grow both in the horizontal (number of base pairs, phylogenomic alignments) as well as vertical (number of taxa) dimension. Put aside the ongoing controversial discussion about appropriate models, partitioning schemes, and assembly methods for phylogenomic alignments, coupled with the high computational cost to infer these, for many organismic groups, a sufficient number of taxa is often exclusively available from one or just a few genes (e.g., rbcL, matK, rDNA). In this paper we address scalability of Maximum-Likelihood-based phylogeny reconstruction with respect to the number of taxa by example of several large nested single-gene rbcL alignments comprising 400 up to 3,491 taxa. In order to thoroughly test the effect of taxon sampling, we deploy an appropriately adapted taxon jackknifing approach. -
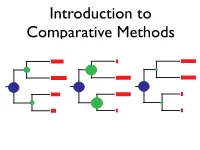
BM CC EB What Can We Learn from a Tree?
Introduction to Comparative Methods BM CC EB What can we learn from a tree? Net diversification (r) Relative extinction (ε) Peridiscaceae Peridiscaceae yllaceae yllaceae h h atop atop Proteaceae Proteaceae r r Ce Ce Tr oc T ho r M M o de c y y H H C C h r r e e a D D a o o nd o e e G G a a m m t t a a d r P r P h h e e u u c c p A p A r e a a e e a a a c a c n n a i B i B h h n d m d l m a l a m m e a e a e e t t n n c u u n n i i d i i e e e e o n o n p n p n a e a S e e S e e n n x x i i r c c a a n o n o p p h g e h g ae e l r a l r a a a a a a i i a a a e a e i b i b h y d c h d c i y i c a a c x c x c c G I a G I a n c n c c c y l y l t a a t a a e e e e e i l c i l c m l m l e c e c f a e a a f a e a a l r r l c c a i i r l e t e t a a r l a a e e u u u u o a o a a a c a c a a l a l e e e b b a a a a e e c e e c a a s c s c c e l c e l e e g e g e a a a a e e n n s e e s e e e e a a a P a P e e N N u u u S u S a e a e a a e e c c l n a l n e e a e e a e a e a e a e r a r a c c C i C i R R a e a e a e a e r c r c A A a d a a d a e i e i phanopetalaceae s r e ph s r e a a s e c s e c e e u u b a a b a e e P P r r l l e n e a a a a m m entho e e e Ha a H o a c r e c r e nt B B e p e e e c e e c a c c h e a p a a p a lo lo l l a a e s o t e s i a r a i a r r r r r a n e a n e a b a l b t a t gaceae e g e ceae a c a s c s a z e z M i a e M i a c a d e a d e ae e ae r e r a e a e a a a c c ce r e r L L i i ac a Vitaceae Vi r r C C e e ta v e v e a a c a a e ea p e ap c a c a e a P P e e l l e Ge G e e ae a t t e e p p r r ce c an u an -

Kenneth J. Wurdack 2,4 and Charles C. Davis
American Journal of Botany 96(8): 1551–1570. 2009. M ALPIGHIALES PHYLOGENETICS: GAINING GROUND ON ONE OF THE MOST RECALCITRANT CLADES IN THE ANGIOSPERM TREE OF LIFE 1 Kenneth J. Wurdack 2,4 and Charles C. Davis3,4 2 Department of Botany, Smithsonian Institution, P.O. Box 37012 NMNH MRC-166, Washington, District of Columbia 20013-7012 USA; and 3 Department of Organismic and Evolutionary Biology, Harvard University Herbaria, 22 Divinity Avenue, Cambridge, Massachusetts 02138 USA The eudicot order Malpighiales contains ~16 000 species and is the most poorly resolved large rosid clade. To clarify phyloge- netic relationships in the order, we used maximum likelihood, Bayesian, and parsimony analyses of DNA sequence data from 13 gene regions, totaling 15 604 bp, and representing all three genomic compartments (i.e., plastid: atpB , matK , ndhF, and rbcL ; mitochondrial: ccmB , cob , matR , nad1B-C , nad6, and rps3; and nuclear: 18S rDNA, PHYC, and newly developed low-copy EMB2765 ). Our sampling of 190 taxa includes representatives from all families of Malpighiales. These data provide greatly in- creased support for the recent additions of Aneulophus , Bhesa , Centroplacus , Ploiarium , and Raffl esiaceae to Malpighiales; sister relations of Phyllanthaceae + Picrodendraceae, monophyly of Hypericaceae, and polyphyly of Clusiaceae. Oxalidales + Huaceae, followed by Celastrales are successive sisters to Malpighiales. Parasitic Raffl esiaceae, which produce the world’ s largest fl owers, are confi rmed as embedded within a paraphyletic Euphorbiaceae. Novel fi ndings show a well-supported placement of Ctenolopho- naceae with Erythroxylaceae + Rhizophoraceae, sister-group relationships of Bhesa + Centroplacus , and the exclusion of Medu- sandra from Malpighiales. New taxonomic circumscriptions include the addition of Bhesa to Centroplacaceae, Medusandra to Peridiscaceae (Saxifragales), Calophyllaceae applied to Clusiaceae subfamily Kielmeyeroideae, Peraceae applied to Euphorbi- aceae subfamily Peroideae, and Huaceae included in Oxalidales. -

Systematics and Biogeography of the Clusioid Clade (Malpighiales) Brad R
Eastern Kentucky University Encompass Biological Sciences Faculty and Staff Research Biological Sciences January 2011 Systematics and Biogeography of the Clusioid Clade (Malpighiales) Brad R. Ruhfel Eastern Kentucky University, [email protected] Follow this and additional works at: http://encompass.eku.edu/bio_fsresearch Part of the Plant Biology Commons Recommended Citation Ruhfel, Brad R., "Systematics and Biogeography of the Clusioid Clade (Malpighiales)" (2011). Biological Sciences Faculty and Staff Research. Paper 3. http://encompass.eku.edu/bio_fsresearch/3 This is brought to you for free and open access by the Biological Sciences at Encompass. It has been accepted for inclusion in Biological Sciences Faculty and Staff Research by an authorized administrator of Encompass. For more information, please contact [email protected]. HARVARD UNIVERSITY Graduate School of Arts and Sciences DISSERTATION ACCEPTANCE CERTIFICATE The undersigned, appointed by the Department of Organismic and Evolutionary Biology have examined a dissertation entitled Systematics and biogeography of the clusioid clade (Malpighiales) presented by Brad R. Ruhfel candidate for the degree of Doctor of Philosophy and hereby certify that it is worthy of acceptance. Signature Typed name: Prof. Charles C. Davis Signature ( ^^^M^ *-^£<& Typed name: Profy^ndrew I^4*ooll Signature / / l^'^ i •*" Typed name: Signature Typed name Signature ^ft/V ^VC^L • Typed name: Prof. Peter Sfe^cnS* Date: 29 April 2011 Systematics and biogeography of the clusioid clade (Malpighiales) A dissertation presented by Brad R. Ruhfel to The Department of Organismic and Evolutionary Biology in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Biology Harvard University Cambridge, Massachusetts May 2011 UMI Number: 3462126 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. -

General View of Malvaceae Juss. S.L. and Taxonomic Revision of Genus Abutilon Mill
JKAU: Sci., Vol. 21 No. 2, pp: 349-363 (2009 A.D. / 1430 A.H.); DOI: 10.4197 / Sci. 21-2.12 General View of Malvaceae Juss. S.L. and Taxonomic Revision of Genus Abutilon Mill. in Saudi Arabia Wafaa Kamal Taia Alexandria University, Faculty of Science, Botany Department, Alexandria, Egypt [email protected] Abstract. This works deals with the recent opinions about the new classification of the core Malvales with special reference to the family Malvaceae s.l. and the morphological description and variations in the species of the genus Abutilon Mill. Taxonomical features of the family as shown in the recent classification systems, with full description of the main divisions of the family. Position of Malvaceae s.l. in the different modern taxonomical systems is clarified. General features of the genus Abutilon stated according to the careful examination of the specimens. Taxonomic position of Abutilon in the Malvaceae is given. Artificial key based on vegetative morphological characters is provided. Keywords: Abutilon, Core Malvales, Eumalvaceae, Morpholog, Systematic Position, Taxonomy. General Features of Family Malvaceae According to Heywood[1] and Watson and Dallwitz[2] the plants of the family Malvaceae s.s. are herbs, shrubs or trees with stipulate, simple, non-sheathing alternate or spiral, petiolate leaves usually with palmate vennation (often three principal veins arising from the base of the leaf blade). Plants are hermaphrodite, rarely dioecious or poly-gamo- monoecious with floral nectarines and entomophilous pollination. Flowers are solitary or aggregating in compound cymes, varying in size from small to large, regular or somewhat irregular, cyclic with distinct calyx and corolla. -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids Xin-Yu Zhu1,2, Mark W Chase3, Yin-Long Qiu4, Hong-Zhi Kong1, David L Dilcher5, Jian-Hua Li6 and Zhi-Duan Chen*1 Address: 1State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, the Chinese Academy of Sciences, Beijing 100093, China, 2Graduate University of the Chinese Academy of Sciences, Beijing 100039, China, 3Jodrell Laboratory, Royal Botanic Gardens, Kew, Richmond, Surrey TW9 3DS, UK, 4Department of Ecology & Evolutionary Biology, The University Herbarium, University of Michigan, Ann Arbor, MI 48108-1048, USA, 5Florida Museum of Natural History, University of Florida, Gainesville, FL 32611-7800, USA and 6Arnold Arboretum of Harvard University, 125 Arborway, Jamaica Plain, MA 02130, USA Email: Xin-Yu Zhu - [email protected]; Mark W Chase - [email protected]; Yin-Long Qiu - [email protected]; Hong- Zhi Kong - [email protected]; David L Dilcher - [email protected]; Jian-Hua Li - [email protected]; Zhi- Duan Chen* - [email protected] * Corresponding author Published: 10 November 2007 Received: 19 June 2007 Accepted: 10 November 2007 BMC Evolutionary Biology 2007, 7:217 doi:10.1186/1471-2148-7-217 This article is available from: http://www.biomedcentral.com/1471-2148/7/217 © 2007 Zhu et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -

Afrostyrax Perkins Gilg from Tropical West Population As a Afrostyrax
I. Anatomical contributions to plant taxonomy I The affinities of Hua Pierre and Afrostyrax Perkins et Gilg P. Baas Rijksherbarium, Leiden Contents Summary 161 Introduction 161 Techniques I <>3 Materials 163 Synopsis of the family Huaceae 163 Results and Discussion 164 Taxonomical and macromorphological notes 164 and seed 165 Ovary, fruit anatomy Pollen morphology t66 167 Vegetative anatomy Hua 167 Afrostyrax 172 The Halphen reaction 179 Discussion of some of the characters of Huaceae 179 of the 182 Hua and Afrostyrax as related genera family Huaceae with other 183 Huaceae compared plant groups Perkins' and Chevalier's view: Styracaceae and related families 183 Hallier's and Hutchinson's suggestions: Linaceae s.l. and Malpighiales 184 Pierre's view: Sterculiaceae and other Malvales 185 General Conclusions *9° Acknowledgements 19° References 191 families Index to original observations in other 192 Summary seed and of and Vegetative anatomy, fruit and structure, pollen morphology Hua Afrostyrax (tropical in anatomical characters West and Central Africa) are described detail. The two genera have many in 182) but are sufficiently different from other families to common (see anatomical family diagnosis on p. discussed and the results of justify the existence of the family Huaceae. Some characteristic features are is comparisons with representatives of about 50 families are reported. Afrostyrax notrelated to Styracaceae, Sterculiaceae and Bombacaceae nor is Hua to Erythroxylaceae as had been suggested in the past. appear to have the highest number ofcharacters incommon with Huaceae of all the families compared. This supports original obser- the inclusion of Huaceae in the Malvales as advocated previously by several authors. -

Antioxidant and Antimicrobial Activities of Two Edibles Spices from Cameroon and Quantification of Their Major Phenolic Compounds
International Food Research Journal 25(6): 2352-2361 (December 2018) Journal homepage: http://www.ifrj.upm.edu.my Antioxidant and antimicrobial activities of two edibles spices from Cameroon and quantification of their major phenolic compounds 1*Sokamte, T. A., 2Mbougueng, P. D., 1Ntsamo B. T. M., 1Noumo, N. T. and 3Tatsadjieu, N. L. 1National School of Agro-Industrial Sciences, Department of Food Science and Nutrition, P.O Box 455, Ngaoundere, Cameroon 2National School of Agro-Industrial Sciences, Department of Process Engineering, University of Ngaoundere, P.O Box 455, Ngaoundere, Cameroon 3University Institute of Technology, Department of Process Engineering and quality control, University of Ngaoundere, P.O Box 454, Ngaoundere, Cameroon Article history Abstract Received: 9 December 2017 This study aimed to analyze the phytochemical composition, antioxidant capacity, antimicrobial Received in revised form: activity and the phenolic profile of Aframomum melegueta and Afrostyrax lepidophyllus 11 February 2018 methanolic extracts. The mean phenolic compound contents were 160.77 ± 6.74 and 46.63 ± Accepted: 20 February 2018 0.93mg GAE/g extract, while that of the tannins was 136.40 ± 0.94 and 8.72 ± 1.60mg GAE/g, respectively for A. melegueta and A. lepidophyllus. Similarly, the flavonoids were higher in A. melegueta extract (70.62 ± 0.90mg CE/g) compared to A. lepidophyllus extract (3.95 ± Keywords 0.14 mg CE/g). A. melegueta extract showed strong antioxidant activity using 2,2-diphenyl- 1-picrylhydrazyl (DPPH), 2,2′-azinobis-3-ethylbenzothiazoline-6-sulfonic acid (ABTS), ferric Antimicrobial activity Antioxidant capacity reducing antioxidant power (FRAP) and Phosphomolybdenum antioxidative power (PAP) Edibles spices tests compared to A. -

2 ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM History Of
ANGIOSPERM PHYLOGENY GROUP (APG) SYSTEM The Angiosperm Phylogeny Group, or APG, refers to an informal international group of systematic botanists who came together to try to establish a consensus view of the taxonomy of flowering plants (angiosperms) that would reflect new knowledge about their relationships based upon phylogenetic studies. As of 2010, three incremental versions of a classification system have resulted from this collaboration (published in 1998, 2003 and 2009). An important motivation for the group was what they viewed as deficiencies in prior angiosperm classifications, which were not based on monophyletic groups (i.e. groups consisting of all the descendants of a common ancestor). APG publications are increasingly influential, with a number of major herbaria changing the arrangement of their collections to match the latest APG system. Angiosperm classification and the APG Until detailed genetic evidence became available, the classification of flowering plants (also known as angiosperms, Angiospermae, Anthophyta or Magnoliophyta) was based on their morphology (particularly that of the flower) and their biochemistry (what kinds of chemical compound they contained or produced). Classification systems were typically produced by an individual botanist or by a small group. The result was a large number of such systems (see List of systems of plant taxonomy). Different systems and their updates tended to be favoured in different countries; e.g. the Engler system in continental Europe; the Bentham & Hooker system in Britain (particularly influential because it was used by Kew); the Takhtajan system in the former Soviet Union and countries within its sphere of influence; and the Cronquist system in the United States. -

The Woody Planet: from Past Triumph to Manmade Decline
plants Review The Woody Planet: From Past Triumph to Manmade Decline Laurence Fazan 1, Yi-Gang Song 2,3 and Gregor Kozlowski 1,3,4,* 1 Department of Biology and Botanical Garden, University of Fribourg, Chemin du Musée 10, 1700 Fribourg, Switzerland; [email protected] 2 Eastern China Conservation Center for Wild Endangered Plant Resources, Shanghai Chenshan Botanical Garden, Chenhua Road No.3888, Songjiang, Shanghai 201602, China; [email protected] 3 Shanghai Chenshan Plant Science Research Center, Chinese Academy of Sciences, Chenhua Road No.3888, Songjiang, Shanghai 201602, China 4 Natural History Museum Fribourg, Chemin du Musée 6, 1700 Fribourg, Switzerland * Correspondence: [email protected]; Tel.: +41-26-300-88-42 Received: 6 November 2020; Accepted: 16 November 2020; Published: 17 November 2020 Abstract: Woodiness evolved in land plants approximately 400 Mya, and very soon after this evolutionary invention, enormous terrestrial surfaces on Earth were covered by dense and luxurious forests. Forests store close to 80% of the biosphere’s biomass, and more than 60% of the global biomass is made of wood (trunks, branches and roots). Among the total number of ca. 374,000 plant species worldwide, approximately 45% (138,500) are woody species—e.g., trees, shrubs or lianas. Furthermore, among all 453 described vascular plant families, 191 are entirely woody (42%). However, recent estimations demonstrate that the woody domination of our planet was even greater before the development of human civilization: 1.4 trillion trees, comprising more than 45% of forest biomass, and 35% of forest cover disappeared during the last few thousands of years of human dominance on our planet.