Phylogenetics, Divergence Times and Diversification from Three Genomic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Partial Flora Survey Rottnest Island Golf Course
PARTIAL FLORA SURVEY ROTTNEST ISLAND GOLF COURSE Prepared by Marion Timms Commencing 1 st Fairway travelling to 2 nd – 11 th left hand side Family Botanical Name Common Name Mimosaceae Acacia rostellifera Summer scented wattle Dasypogonaceae Acanthocarpus preissii Prickle lily Apocynaceae Alyxia Buxifolia Dysentry bush Casuarinacea Casuarina obesa Swamp sheoak Cupressaceae Callitris preissii Rottnest Is. Pine Chenopodiaceae Halosarcia indica supsp. Bidens Chenopodiaceae Sarcocornia blackiana Samphire Chenopodiaceae Threlkeldia diffusa Coast bonefruit Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Suada australis Seablite Chenopodiaceae Atriplex isatidea Coast saltbush Poaceae Sporabolis virginicus Marine couch Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Pittosporaceae Pittosporum phylliraeoides Weeping pittosporum Poaceae Stipa flavescens Tussock grass 2nd – 11 th Fairway Family Botanical Name Common Name Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Atriplex isatidea Coast saltbush Cyperaceae Gahnia trifida Coast sword sedge Pittosporaceae Pittosporum phyliraeoides Weeping pittosporum Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Chenopodiaceae Sarcocornia blackiana Samphire Central drainage wetland commencing at Vietnam sign Family Botanical Name Common Name Chenopodiaceae Halosarcia halecnomoides Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Sarcocornia blackiana Samphire Poaceae Sporobolis virginicus Cyperaceae Gahnia Trifida Coast sword sedge -

Restoration After Removal of Pines at Gnangara Final
RESTORATION OF BANKSIA WOODLAND AFTER THE REMOVAL OF PINES AT GNANGARA: SEED SPECIES REQUIREMENTS AND PRESCRIPTIONS FOR RESTORATION A report prepared on behalf of the Department of Environment and Conservation for the Gnangara Sustainability Strategy Kellie Maher University of Western Australia May 2009 Restoration of Banksia woodland after the removal of pines at Gnangara: seed species requirements and prescriptions for restoration Report for the Department of Environment and Conservation Kellie Maher University of Western Australia Gnangara Sustainability Strategy Taskforce Department of Water 168 St Georges Terrace Perth Western Australia 6000 Telephone +61 8 6364 7600 Facsimile +61 8 6364 7601 www.gnangara.water.wa.gov.au © Government of Western Australia 2009 May 2009 This work is copyright. You may download, display, print and reproduce this material in unaltered form only (retaining this notice) for your personal, non-commercial use or use within your organisation. Apart from any use as permitted under the Copyright Act 1968 , all other rights are reserved. Requests and inquiries concerning reproduction and rights should be addressed to the Department of Conservation and Environment. This document has been commissioned/produced as part of the Gnangara Sustainability Strategy (GSS). The GSS is a State Government initiative which aims to provide a framework for a whole of government approach to address land use and water planning issues associated with the Gnangara groundwater system. For more information go to www.gnangara.water.wa.gov.au 1 Restoration of Banksia woodland after the removal of pines at Gnangara: seed species requirements and prescriptions for restoration A report to the Department of Environment and Conservation Kellie Maher University of Western Australia May 2009 2 Table of Contents List of Tables .................................................................................................................... -

Complete Chloroplast Genomes Shed Light on Phylogenetic
www.nature.com/scientificreports OPEN Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae) Ju Namgung1,4, Hoang Dang Khoa Do1,2,4, Changkyun Kim1, Hyeok Jae Choi3 & Joo‑Hwan Kim1* Allioideae includes economically important bulb crops such as garlic, onion, leeks, and some ornamental plants in Amaryllidaceae. Here, we reported the complete chloroplast genome (cpDNA) sequences of 17 species of Allioideae, fve of Amaryllidoideae, and one of Agapanthoideae. These cpDNA sequences represent 80 protein‑coding, 30 tRNA, and four rRNA genes, and range from 151,808 to 159,998 bp in length. Loss and pseudogenization of multiple genes (i.e., rps2, infA, and rpl22) appear to have occurred multiple times during the evolution of Alloideae. Additionally, eight mutation hotspots, including rps15-ycf1, rps16-trnQ-UUG, petG-trnW-CCA , psbA upstream, rpl32- trnL-UAG , ycf1, rpl22, matK, and ndhF, were identifed in the studied Allium species. Additionally, we present the frst phylogenomic analysis among the four tribes of Allioideae based on 74 cpDNA coding regions of 21 species of Allioideae, fve species of Amaryllidoideae, one species of Agapanthoideae, and fve species representing selected members of Asparagales. Our molecular phylogenomic results strongly support the monophyly of Allioideae, which is sister to Amaryllioideae. Within Allioideae, Tulbaghieae was sister to Gilliesieae‑Leucocoryneae whereas Allieae was sister to the clade of Tulbaghieae‑ Gilliesieae‑Leucocoryneae. Molecular dating analyses revealed the crown age of Allioideae in the Eocene (40.1 mya) followed by diferentiation of Allieae in the early Miocene (21.3 mya). The split of Gilliesieae from Leucocoryneae was estimated at 16.5 mya. -
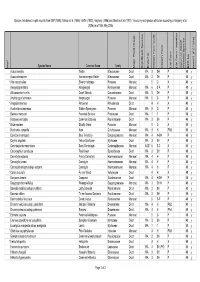
BFS048 Site Species List
Species lists based on plot records from DEP (1996), Gibson et al. (1994), Griffin (1993), Keighery (1996) and Weston et al. (1992). Taxonomy and species attributes according to Keighery et al. (2006) as of 16th May 2005. Species Name Common Name Family Major Plant Group Significant Species Endemic Growth Form Code Growth Form Life Form Life Form - aquatics Common SSCP Wetland Species BFS No kens01 (FCT23a) Wd? Acacia sessilis Wattle Mimosaceae Dicot WA 3 SH P 48 y Acacia stenoptera Narrow-winged Wattle Mimosaceae Dicot WA 3 SH P 48 y * Aira caryophyllea Silvery Hairgrass Poaceae Monocot 5 G A 48 y Alexgeorgea nitens Alexgeorgea Restionaceae Monocot WA 6 S-R P 48 y Allocasuarina humilis Dwarf Sheoak Casuarinaceae Dicot WA 3 SH P 48 y Amphipogon turbinatus Amphipogon Poaceae Monocot WA 5 G P 48 y * Anagallis arvensis Pimpernel Primulaceae Dicot 4 H A 48 y Austrostipa compressa Golden Speargrass Poaceae Monocot WA 5 G P 48 y Banksia menziesii Firewood Banksia Proteaceae Dicot WA 1 T P 48 y Bossiaea eriocarpa Common Bossiaea Papilionaceae Dicot WA 3 SH P 48 y * Briza maxima Blowfly Grass Poaceae Monocot 5 G A 48 y Burchardia congesta Kara Colchicaceae Monocot WA 4 H PAB 48 y Calectasia narragara Blue Tinsel Lily Dasypogonaceae Monocot WA 4 H-SH P 48 y Calytrix angulata Yellow Starflower Myrtaceae Dicot WA 3 SH P 48 y Centrolepis drummondiana Sand Centrolepis Centrolepidaceae Monocot AUST 6 S-C A 48 y Conostephium pendulum Pearlflower Epacridaceae Dicot WA 3 SH P 48 y Conostylis aculeata Prickly Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis juncea Conostylis Haemodoraceae Monocot WA 4 H P 48 y Conostylis setigera subsp. -

Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University
Aliso: A Journal of Systematic and Evolutionary Botany Volume 22 | Issue 1 Article 3 2006 Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis Kåre Bremer Uppsala University Thomas Janssen Muséum National d'Histoire Naturelle Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons Recommended Citation Bremer, Kåre and Janssen, Thomas (2006) "Gondwanan Origin of Major Monocot Groups Inferred from Dispersal-Vicariance Analysis," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 22: Iss. 1, Article 3. Available at: http://scholarship.claremont.edu/aliso/vol22/iss1/3 Aliso 22, pp. 22-27 © 2006, Rancho Santa Ana Botanic Garden GONDWANAN ORIGIN OF MAJOR MONO COT GROUPS INFERRED FROM DISPERSAL-VICARIANCE ANALYSIS KARE BREMERl.3 AND THOMAS JANSSEN2 lDepartment of Systematic Botany, Evolutionary Biology Centre, Norbyvagen l8D, SE-752 36 Uppsala, Sweden; 2Museum National d'Histoire Naturelle, Departement de Systematique et Evolution, USM 0602: Taxonomie et collections, 16 rue Buffon, 75005 Paris, France 3Corresponding author ([email protected]) ABSTRACT Historical biogeography of major monocot groups was investigated by biogeographical analysis of a dated phylogeny including 79 of the 81 monocot families using the Angiosperm Phylogeny Group II (APG II) classification. Five major areas were used to describe the family distributions: Eurasia, North America, South America, Africa including Madagascar, and Australasia including New Guinea, New Caledonia, and New Zealand. In order to investigate the possible correspondence with continental breakup, the tree with its terminal distributions was fitted to the geological area cladogram «Eurasia, North America), (Africa, (South America, Australasia») and to alternative area cladograms using the TreeFitter program. -

Download the Full Report Pdf, 2.9 MB
VKM Report 2016:50 Assessment of the risks to Norwegian biodiversity from the import and keeping of aquarium and garden pond plants Opinion of the Panel on Alien Organisms and Trade in Endangered Species (CITES) of the Norwegian Scientific Committee for Food Safety Report from the Norwegian Scientific Committee for Food Safety (VKM) 2016:50 Assessment of the risks to Norwegian biodiversity from the import and keeping of aquarium and garden pond plants Opinion of the Panel on Alien Organisms and Trade in Endangered Species (CITES) of the Norwegian Scientific Committee for Food Safety 01.11.2016 ISBN: 00000-00000 Norwegian Scientific Committee for Food Safety (VKM) Po 4404 Nydalen N – 0403 Oslo Norway Phone: +47 21 62 28 00 Email: [email protected] www.vkm.no www.english.vkm.no Suggested citation: VKM (2016). Assessment of the risks to Norwegian biodiversity from the import and keeping of aquarium and garden pond plants. Scientific Opinion on the on Alien Organisms and Trade in Endangered species of the Norwegian Scientific Committee for Food Safety ISBN: 978-82-8259-240-6, Oslo, Norway. VKM Report 2016:50 Title: Assessment of the risks to Norwegian biodiversity from the import and keeping of aquarium and garden pond plants Authors preparing the draft opinion Hugo de Boer (chair), Maria G. Asmyhr (VKM staff), Hanne H. Grundt, Inga Kjersti Sjøtun, Hans K. Stenøien, Iris Stiers. Assessed and approved The opinion has been assessed and approved by Panel on Alien organisms and Trade in Endangered Species (CITES). Members of the panel are: Vigdis Vandvik (chair), Hugo de Boer, Jan Ove Gjershaug, Kjetil Hindar, Lawrence Kirkendall, Nina Elisabeth Nagy, Anders Nielsen, Eli K. -

Winter 2004.Pmd
The Lady-Slipper, 19:4 / Winter 2004 1 The Lady-Slipper Kentucky Native Plant Society Number 19:4 Winter 2004 A Message from the President: It’s Membership Winter is upon us. I hope everyone had some opportunity to experience the colors of Fall and now some of us will turn our attention to winter botany. While I was unable to Renewal Time! attend, I understand that our Fall meeting at Shakertown Kentucky Native Plant Society with Dr. Bill Bryant from Thomas More College as the guest EMBERSHIP ORM speaker was a great success. M F Our Native Plant Certification program was relatively successful this Fall. Plant taxonomy failed to meet Name(s) ____________________________________ because the NKU’s Community Education Bulletin was Address ____________________________________ mailed too late for anyone to sign up for the course. The woody plants course did, however, have a successful run. City, State, Zip ______________________________ This coming Spring, we will be offering Basic Plant Taxonomy, Plant Communities and Spring Wildflowers of KY County __________________________________ Central Kentucky. Tel.: (home) ______________________________ You will see in this issue that we are promoting “Chinquapin” the newsletter of the Southern Appalachian (work) ______________________________ Botanical Society (SABS). SABS is an organization E-mail _______________________________ largely made up of professional botanists and produces a quarterly scholarly journal. The newsletter “Chinquapin” o Add me to the e-mail list for time-critical native plant news has more of a general interest approach much like our o Include my contact info in any future KNPS Member Directory newsletter but on a regional scale. In this issue we have Membership Categories: provided subscription information on page 7. -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M. -

GENOME EVOLUTION in MONOCOTS a Dissertation
GENOME EVOLUTION IN MONOCOTS A Dissertation Presented to The Faculty of the Graduate School At the University of Missouri In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy By Kate L. Hertweck Dr. J. Chris Pires, Dissertation Advisor JULY 2011 The undersigned, appointed by the dean of the Graduate School, have examined the dissertation entitled GENOME EVOLUTION IN MONOCOTS Presented by Kate L. Hertweck A candidate for the degree of Doctor of Philosophy And hereby certify that, in their opinion, it is worthy of acceptance. Dr. J. Chris Pires Dr. Lori Eggert Dr. Candace Galen Dr. Rose‐Marie Muzika ACKNOWLEDGEMENTS I am indebted to many people for their assistance during the course of my graduate education. I would not have derived such a keen understanding of the learning process without the tutelage of Dr. Sandi Abell. Members of the Pires lab provided prolific support in improving lab techniques, computational analysis, greenhouse maintenance, and writing support. Team Monocot, including Dr. Mike Kinney, Dr. Roxi Steele, and Erica Wheeler were particularly helpful, but other lab members working on Brassicaceae (Dr. Zhiyong Xiong, Dr. Maqsood Rehman, Pat Edger, Tatiana Arias, Dustin Mayfield) all provided vital support as well. I am also grateful for the support of a high school student, Cady Anderson, and an undergraduate, Tori Docktor, for their assistance in laboratory procedures. Many people, scientist and otherwise, helped with field collections: Dr. Travis Columbus, Hester Bell, Doug and Judy McGoon, Julie Ketner, Katy Klymus, and William Alexander. Many thanks to Barb Sonderman for taking care of my greenhouse collection of many odd plants brought back from the field. -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

The Complete Chloroplast Genome Sequence of Asparagus (Asparagus Officinalis L.) and Its Phy- Logenetic Positon Within Asparagales
Central International Journal of Plant Biology & Research Bringing Excellence in Open Access Research Note *Corresponding author Wentao Sheng, Department of Biological Technology, Nanchang Normal University, Nanchang 330032, The Complete Chloroplast Jiangxi, China, Tel: 86-0791-87619332; Fax: 86-0791- 87619332; Email: Submitted: 14 September 2017 Genome Sequence of Accepted: 09 October 2017 Published: 10 October 2017 Asparagus (Asparagus ISSN: 2333-6668 Copyright © 2017 Sheng et al. officinalis L.) and its OPEN ACCESS Keywords Phylogenetic Positon within • Asparagus officinalis L • Chloroplast genome • Phylogenomic evolution Asparagales • Asparagales Wentao Sheng*, Xuewen Chai, Yousheng Rao, Xutang, Tu, and Shangguang Du Department of Biological Technology, Nanchang Normal University, China Abstract Asparagus (Asparagus officinalis L.) is a horticultural homology of medicine and food with health care. The entire chloroplast (cp) genome of asparagus was sequenced with Hiseq4000 platform. The complete cp genome maps a circular molecule of 156,699bp built with a quadripartite organization: two inverted repeats (IRs) of 26,531bp, separated by a large single copy (LSC) sequence of 84,999bp and a small single copy (SSC) sequence of 18,638bp. A total of 112 genes comprising of 78 protein-coding genes, 30 tRNAs and 4 rRNAs were successfully annotated, 17 of which included introns. The identity, number and GC content of asparagus cp genes were similar to those of other asparagus species genomes. Analysis revealed 81 simple sequence repeat (SSR) loci, most composed of A or T, contributing to a bias in base composition. A maximum likelihood phylogenomic evolution analysis showed that asparagus was closely related to Polygonatum cyrtonema that belonged to the genus Asparagales. -

TELOPEA Publication Date: 13 October 1983 Til
Volume 2(4): 425–452 TELOPEA Publication Date: 13 October 1983 Til. Ro)'al BOTANIC GARDENS dx.doi.org/10.7751/telopea19834408 Journal of Plant Systematics 6 DOPII(liPi Tmst plantnet.rbgsyd.nsw.gov.au/Telopea • escholarship.usyd.edu.au/journals/index.php/TEL· ISSN 0312-9764 (Print) • ISSN 2200-4025 (Online) Telopea 2(4): 425-452, Fig. 1 (1983) 425 CURRENT ANATOMICAL RESEARCH IN LILIACEAE, AMARYLLIDACEAE AND IRIDACEAE* D.F. CUTLER AND MARY GREGORY (Accepted for publication 20.9.1982) ABSTRACT Cutler, D.F. and Gregory, Mary (Jodrell(Jodrel/ Laboratory, Royal Botanic Gardens, Kew, Richmond, Surrey, England) 1983. Current anatomical research in Liliaceae, Amaryllidaceae and Iridaceae. Telopea 2(4): 425-452, Fig.1-An annotated bibliography is presented covering literature over the period 1968 to date. Recent research is described and areas of future work are discussed. INTRODUCTION In this article, the literature for the past twelve or so years is recorded on the anatomy of Liliaceae, AmarylIidaceae and Iridaceae and the smaller, related families, Alliaceae, Haemodoraceae, Hypoxidaceae, Ruscaceae, Smilacaceae and Trilliaceae. Subjects covered range from embryology, vegetative and floral anatomy to seed anatomy. A format is used in which references are arranged alphabetically, numbered and annotated, so that the reader can rapidly obtain an idea of the range and contents of papers on subjects of particular interest to him. The main research trends have been identified, classified, and check lists compiled for the major headings. Current systematic anatomy on the 'Anatomy of the Monocotyledons' series is reported. Comment is made on areas of research which might prove to be of future significance.