WWOX Loses the Ability to Regulate Oncogenic AP-2Γ and Synergizes with Tumor Suppressor AP-2Α in High-Grade Bladder Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cellular Responses to Erbb-2 Overexpression in Human Mammary Luminal Epithelial Cells: Comparison of Mrna and Protein Expression
British Journal of Cancer (2004) 90, 173 – 181 & 2004 Cancer Research UK All rights reserved 0007 – 0920/04 $25.00 www.bjcancer.com Cellular responses to ErbB-2 overexpression in human mammary luminal epithelial cells: comparison of mRNA and protein expression SL White1, S Gharbi1, MF Bertani1, H-L Chan1, MD Waterfield1 and JF Timms*,1 1 Ludwig Institute for Cancer Research, Wing 1.1, Cruciform Building, Gower Street, London WCIE 6BT, UK Microarray analysis offers a powerful tool for studying the mechanisms of cellular transformation, although the correlation between mRNA and protein expression is largely unknown. In this study, a microarray analysis was performed to compare transcription in response to overexpression of the ErbB-2 receptor tyrosine kinase in a model mammary luminal epithelial cell system, and in response to the ErbB-specific growth factor heregulin b1. We sought to validate mRNA changes by monitoring changes at the protein level using a parallel proteomics strategy, and report a surprisingly high correlation between transcription and translation for the subset of genes studied. We further characterised the identified targets and relate differential expression to changes in the biological properties of ErbB-2-overexpressing cells. We found differential regulation of several key cell cycle modulators, including cyclin D2, and downregulation of a large number of interferon-inducible genes, consistent with increased proliferation of the ErbB-2- overexpressing cells. Furthermore, differential expression of genes involved in extracellular matrix modelling and cellular adhesion was linked to altered adhesion of these cells. Finally, we provide evidence for enhanced autocrine activation of MAPK signalling and the AP-1 transcription complex. -

Reconstruction of the Global Neural Crest Gene Regulatory Network in Vivo
Reconstruction of the global neural crest gene regulatory network in vivo Ruth M Williams1, Ivan Candido-Ferreira1, Emmanouela Repapi2, Daria Gavriouchkina1,4, Upeka Senanayake1, Jelena Telenius2,3, Stephen Taylor2, Jim Hughes2,3, and Tatjana Sauka-Spengler1,∗ Supplemental Material ∗Lead and corresponding author: Tatjana Sauka-Spengler ([email protected]) 1University of Oxford, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, Oxford, OX3 9DS, UK 2University of Oxford, MRC Centre for Computational Biology, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 3University of Oxford, MRC Molecular Haematology Unit, MRC Weatherall Institute of Molecular Medicine, Oxford, OX3 9DS, UK 4Present Address: Okinawa Institute of Science and Technology, Molecular Genetics Unit, Onna, 904-0495, Japan A 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R1_5-6ss) log2(R1_8-10ss) log2(R1_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.92 0 r=0.99 0 r=0.96 0 r=0.99 0 r=0.96 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R2_non-NC) log2(R2_5-6ss) log2(R3_5-6ss) log2(R2_8-10ss) log2(R3_8-10ss) 25 25 25 25 25 20 20 20 20 20 15 15 15 15 15 10 10 10 10 10 log2(R1_5-6ss) log2(R2_5-6ss) log2(R1_8-10ss) log2(R2_8-10ss) log2(R1_non-NC) 5 5 5 5 5 0 r=0.94 0 r=0.96 0 r=0.95 0 r=0.96 0 r=0.95 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 0 5 10 15 20 25 log2(R3_non-NC) log2(R4_5-6ss) log2(R3_5-6ss) log2(R4_8-10ss) log2(R3_8-10ss) -

Gene PMID WBS Locus ABR 26603386 AASDH 26603386
Supplementary material J Med Genet Gene PMID WBS Locus ABR 26603386 AASDH 26603386 ABCA1 21304579 ABCA13 26603386 ABCA3 25501393 ABCA7 25501393 ABCC1 25501393 ABCC3 25501393 ABCG1 25501393 ABHD10 21304579 ABHD11 25501393 yes ABHD2 25501393 ABHD5 21304579 ABLIM1 21304579;26603386 ACOT12 25501393 ACSF2,CHAD 26603386 ACSL4 21304579 ACSM3 26603386 ACTA2 25501393 ACTN1 26603386 ACTN3 26603386;25501393;25501393 ACTN4 21304579 ACTR1B 21304579 ACVR2A 21304579 ACY3 19897463 ACYP1 21304579 ADA 25501393 ADAM12 21304579 ADAM19 25501393 ADAM32 26603386 ADAMTS1 25501393 ADAMTS10 25501393 ADAMTS12 26603386 ADAMTS17 26603386 ADAMTS6 21304579 ADAMTS7 25501393 ADAMTSL1 21304579 ADAMTSL4 25501393 ADAMTSL5 25501393 ADCY3 25501393 ADK 21304579 ADRBK2 25501393 AEBP1 25501393 AES 25501393 AFAP1,LOC84740 26603386 AFAP1L2 26603386 AFG3L1 21304579 AGAP1 26603386 AGAP9 21304579 Codina-Sola M, et al. J Med Genet 2019; 56:801–808. doi: 10.1136/jmedgenet-2019-106080 Supplementary material J Med Genet AGBL5 21304579 AGPAT3 19897463;25501393 AGRN 25501393 AGXT2L2 25501393 AHCY 25501393 AHDC1 26603386 AHNAK 26603386 AHRR 26603386 AIDA 25501393 AIFM2 21304579 AIG1 21304579 AIP 21304579 AK5 21304579 AKAP1 25501393 AKAP6 21304579 AKNA 21304579 AKR1E2 26603386 AKR7A2 21304579 AKR7A3 26603386 AKR7L 26603386 AKT3 21304579 ALDH18A1 25501393;25501393 ALDH1A3 21304579 ALDH1B1 21304579 ALDH6A1 21304579 ALDOC 21304579 ALG10B 26603386 ALG13 21304579 ALKBH7 25501393 ALPK2 21304579 AMPH 21304579 ANG 21304579 ANGPTL2,RALGPS1 26603386 ANGPTL6 26603386 ANK2 21304579 ANKMY1 26603386 ANKMY2 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supporting Information
Supporting Information Edgar et al. 10.1073/pnas.1601895113 SI Methods (Actimetrics), and recordings were analyzed using LumiCycle Mice. Sample size was determined using the resource equation: Data Analysis software (Actimetrics). E (degrees of freedom in ANOVA) = (total number of exper- – Cell Cycle Analysis of Confluent Cell Monolayers. NIH 3T3, primary imental animals) (number of experimental groups), with −/− sample size adhering to the condition 10 < E < 20. For com- WT, and Bmal1 fibroblasts were sequentially transduced − − parison of MuHV-4 and HSV-1 infection in WT vs. Bmal1 / with lentiviral fluorescent ubiquitin-based cell cycle indicators mice at ZT7 (Fig. 2), the investigator did not know the genotype (FUCCI) mCherry::Cdt1 and amCyan::Geminin reporters (32). of the animals when conducting infections, bioluminescence Dual reporter-positive cells were selected by FACS (Influx Cell imaging, and quantification. For bioluminescence imaging, Sorter; BD Biosciences) and seeded onto 35-mm dishes for mice were injected intraperitoneally with endotoxin-free lucif- subsequent analysis. To confirm that expression of mCherry:: Cdt1 and amCyan::Geminin correspond to G1 (2n DNA con- erin (Promega E6552) using 2 mg total per mouse. Following < ≤ anesthesia with isofluorane, they were scanned with an IVIS tent) and S/G2 (2 n 4 DNA content) cell cycle phases, Lumina (Caliper Life Sciences), 15 min after luciferin admin- respectively, cells were stained with DNA dye DRAQ5 (abcam) and analyzed by flow cytometry (LSR-Fortessa; BD Biosci- istration. Signal intensity was quantified using Living Image ences). To examine dynamics of replicative activity under ex- software (Caliper Life Sciences), obtaining maximum radiance perimental confluent conditions, synchronized FUCCI reporter for designated regions of interest (photons per second per − − − monolayers were observed by time-lapse live cell imaging over square centimeter per Steradian: photons·s 1·cm 2·sr 1), relative 3 d (Nikon Eclipse Ti-E inverted epifluorescent microscope). -
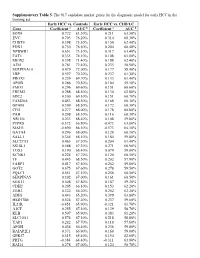
Supplementary Table 5. the 917 Candidate Marker Genes for the Diagnostic Model for Early HCC in the Training Set
Supplementary Table 5. The 917 candidate marker genes for the diagnostic model for early HCC in the training set. Early HCC vs. Controls Early HCC vs. CHB/LC Gene Coefficient a AUC b Coefficient a AUC b SOX9 0.722 81.30% 0.211 63.50% EVC 0.703 76.20% 0.314 68.30% CHST9 0.398 75.50% 0.150 62.40% PDX1 0.730 76.50% 0.204 60.40% NPBWR1 0.651 73.10% 0.317 63.40% FAT1 0.335 74.10% 0.108 61.00% MEIS2 0.398 71.40% 0.188 62.40% A2M 0.761 72.40% 0.235 58.90% SERPINA10 0.479 72.00% 0.177 58.40% LBP 0.597 70.20% 0.237 61.30% PROX1 0.239 69.70% 0.133 61.40% APOB 0.286 70.50% 0.104 59.10% FMO3 0.296 69.60% 0.151 60.60% FREM2 0.288 68.50% 0.130 62.80% SDC2 0.300 69.10% 0.151 60.70% FAM20A 0.453 68.50% 0.168 60.30% GPAM 0.309 68.50% 0.172 60.30% CFH 0.277 68.00% 0.178 60.80% PAH 0.208 68.30% 0.116 60.30% NR1H4 0.233 68.40% 0.108 59.80% PTPRS -0.572 66.80% -0.473 63.00% SIAH3 -0.690 66.10% -0.573 64.10% GATA4 0.296 68.00% 0.128 60.10% SALL1 0.344 68.10% 0.184 59.80% SLC27A5 0.463 67.30% 0.204 61.40% SS18L1 0.588 67.30% 0.271 60.90% TOX3 0.190 68.40% 0.079 59.00% KCNK1 0.224 67.70% 0.120 60.10% TF 0.445 68.50% 0.202 57.90% FARP1 0.417 67.50% 0.252 59.80% GOT2 0.675 67.60% 0.278 59.50% PQLC1 0.651 67.10% 0.258 60.50% SERPINA5 0.302 67.00% 0.161 60.50% SOX13 0.508 67.80% 0.187 59.30% CDH2 0.205 66.10% 0.153 62.20% ITIH2 0.322 66.20% 0.252 62.20% ADIG 0.443 65.20% 0.399 63.80% HSD17B6 0.524 67.20% 0.237 59.60% IL21R -0.451 65.90% -0.321 61.70% A1CF 0.255 67.10% 0.139 58.70% KLB 0.507 65.90% 0.383 61.20% SLC10A1 0.574 67.10% 0.218 58.80% YAP1 0.282 67.70% 0.118 -

Systematic Detection of Divergent Brain Protein-Coding Genes in Human Evolution
bioRxiv preprint doi: https://doi.org/10.1101/658658; this version posted January 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Title: Systematic detection of divergent brain protein-coding genes in human evolution 2 and their roles in cognition 3 Short title: Divergent brain protein-coding genes in human evolution 4 5 Guillaume Dumasa, Simon Malesysa and Thomas Bourgerona 6 a Human Genetics and Cognitive Functions, Institut Pasteur, UMR3571 CNRS, Université de Paris, Paris, 7 (75015) France 8 9 10 Corresponding author: 11 Guillaume Dumas 12 Human Genetics and Cognitive Functions 13 Institut Pasteur 14 75015 Paris, France 15 Phone: +33 6 28 25 56 65 16 [email protected] Dumas, Malesys and Bourgeron 1 of 37 bioRxiv preprint doi: https://doi.org/10.1101/658658; this version posted January 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 17 Abstract 18 The human brain differs from that of other primates, but the genetic basis of these differences 19 remains unclear. We investigated the evolutionary pressures acting on almost all human 20 protein-coding genes (N=11,667; 1:1 orthologs in primates) on the basis of their divergence 21 from those of early hominins, such as Neanderthals, and non-human primates. -

Integration of 198 Chip-Seq Datasets Reveals Human Cis-Regulatory Regions
Integration of 198 ChIP-seq datasets reveals human cis-regulatory regions Hamid Bolouri & Larry Ruzzo Thanks to Stephen Tapscott, Steven Henikoff & Zizhen Yao Slides will be available from: http://labs.fhcrc.org/bolouri/ Email [email protected] for manuscript (Bolouri & Ruzzo, submitted) Kleinjan & van Heyningen, Am. J. Hum. Genet., 2005, (76)8–32 Epstein, Briefings in Func. Genom. & Protemoics, 2009, 8(4)310-16 Regulation of SPi1 (Sfpi1, PU.1 protein) expression – part 1 miR155*, miR569# ~750nt promoter ~250nt promoter The antisense RNA • causes translational stalling • has its own promoter • requires distal SPI1 enhancer • is transcribed with/without SPI1. # Hikami et al, Arthritis & Rheumatism, 2011, 63(3):755–763 * Vigorito et al, 2007, Immunity 27, 847–859 Ebralidze et al, Genes & Development, 2008, 22: 2085-2092. Regulation of SPi1 expression – part 2 (mouse coordinates) Bidirectional ncRNA transcription proportional to PU.1 expression PU.1/ELF1/FLI1/GLI1 GATA1 GATA1 Sox4/TCF/LEF PU.1 RUNX1 SP1 RUNX1 RUNX1 SP1 ELF1 NF-kB SATB1 IKAROS PU.1 cJun/CEBP OCT1 cJun/CEBP 500b 500b 500b 500b 500b 750b 500b -18Kb -14Kb -12Kb -10Kb -9Kb Chou et al, Blood, 2009, 114: 983-994 Hoogenkamp et al, Molecular & Cellular Biology, 2007, 27(21):7425-7438 Zarnegar & Rothenberg, 2010, Mol. & cell Biol. 4922-4939 An NF-kB binding-site variant in the SPI1 URE reduces PU.1 expression & is GGGCCTCCCC correlated with AML GGGTCTTCCC Bonadies et al, Oncogene, 2009, 29(7):1062-72. SATB1 binding site A distal single nucleotide polymorphism alters long- range regulation of the PU.1 gene in acute myeloid leukemia Steidl et al, J Clin Invest. -

Transcriptional and Post-Transcriptional Regulation of ATP-Binding Cassette Transporter Expression
Transcriptional and Post-transcriptional Regulation of ATP-binding Cassette Transporter Expression by Aparna Chhibber DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Pharmaceutical Sciences and Pbarmacogenomies in the Copyright 2014 by Aparna Chhibber ii Acknowledgements First and foremost, I would like to thank my advisor, Dr. Deanna Kroetz. More than just a research advisor, Deanna has clearly made it a priority to guide her students to become better scientists, and I am grateful for the countless hours she has spent editing papers, developing presentations, discussing research, and so much more. I would not have made it this far without her support and guidance. My thesis committee has provided valuable advice through the years. Dr. Nadav Ahituv in particular has been a source of support from my first year in the graduate program as my academic advisor, qualifying exam committee chair, and finally thesis committee member. Dr. Kathy Giacomini graciously stepped in as a member of my thesis committee in my 3rd year, and Dr. Steven Brenner provided valuable input as thesis committee member in my 2nd year. My labmates over the past five years have been incredible colleagues and friends. Dr. Svetlana Markova first welcomed me into the lab and taught me numerous laboratory techniques, and has always been willing to act as a sounding board. Michael Martin has been my partner-in-crime in the lab from the beginning, and has made my days in lab fly by. Dr. Yingmei Lui has made the lab run smoothly, and has always been willing to jump in to help me at a moment’s notice. -

A Multi-Exon Deletion Within WWOX Is Associated with a 46,XY Disorder of Sex
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Ghent University Academic Bibliography ͳ A multi-exon deletion within WWOX is associated with a 46,XY Disorder of Sex ʹ Development. ͵ Ͷ Stefan White1,2, Jacqueline Hewitt1,3, Erin Turbitt1, Yvonne van der Zwan4, RemKo ͷ Hersmus4, Sten Drop4, Peter Koopman5, Vincent Harley6, Martine Cools7, Leendert Looijenga4, Andrew Sinclair1,3 ͺ 1. Murdoch Childrens Research Institute, Royal Childrens Hospital, Melbourne, VIC, ͻ Australia ͳͲ 2. Monash Institute of Medical Research, Melbourne, VIC, Australia ͳͳ 3. The University of Melbourne, Melbourne, VIC, Australia ͳʹ 4. Erasmus MC-University Medical Center, Rotterdam, The Netherlands ͳ͵ 5. Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, ͳͶ Australia ͳͷ 6. Prince Henrys Institute, Melbourne, VIC, Australia ͳ 7. University Hospital Ghent, Ghent, Belgium ͳ ͳͺ Corresponding author: ͳͻ Dr Stefan White ʹͲ Monash Institute of Medical Research ʹͳ 27-31 Wright Street ʹʹ Clayton VIC 3168 ʹ͵ AUSTRALIA ʹͶ Email [email protected] ʹͷ Tel +61 3 9902 4812 ʹ Fax +61 3 9594 7114 ʹ Running title: WWOX and Disorders of Sex Development ʹͺ ʹͻ Abstract ͵Ͳ Disorders of sex development (DSD) are congenital conditions where chromosomal, ͵ͳ gonad or genital development is atypical. In a significant proportion of 46,XY DSD ͵ʹ cases it is not possible to identify a causative mutation, maKing genetic counseling ͵͵ difficult and potentially hindering optimal treatment. We present here the analysis of a ͵Ͷ 46,XY DSD patient that presented at birth with ambiguous genitalia. Histological ͵ͷ analysis of the surgically-removed gonads showed bilateral undifferentiated gonadal ͵ tissue and immature testis, both containing malignant germ cells. -

1 Characterization of Wwox Expression and Function in Canine
Characterization of Wwox Expression and Function in Canine Mast Cell Tumors and Malignant Mast Cell Lines THESIS Presented in Partial Fulfillment of the Requirements for the Degree Master of Science in the Graduate School of The Ohio State University By Rebecca Lynn Makii Graduate Program in Comparative and Veterinary Medicine The Ohio State University 2020 Master’s Examination Committee Joelle Fenger, DVM, PhD, DACVIM, Advisor Eric Green, DVM, DACVR Ryan Jennings, DVM, PhD, DACVP 1 Copyrighted by Rebecca Lynn Makii 2020 2 Abstract Mast cell tumors (MCT) are the most common skin tumor in dogs with behavior varying from benign to aggressive, metastatic disease. While activating mutations in the receptor tyrosine kinase KIT (c-KIT) have been identified in up to 30% of high-grade MCTs, the genetic alterations driving tumorigenesis in the 70% of MCTs that do not possess c-KIT mutations remains unclear. The WW domain-containing oxidoreductase (WWOX) tumor suppressor gene is frequently lost or attenuated in many human cancers and cancer cell lines and data suggest that loss of WWOX impedes DNA damage response (DDR) and repair leading to genomic instability. The purpose of this study was to characterize WWOX expression in spontaneous canine mast MCTs and mastocytoma cell lines and begin to define the functional consequences of WWOX inactivation on mast cell viability and clonogenic survival in response to double-stranded DNA (dsDNA) damaging agents. qRT-PCR and Western blotting showed that WWOX is decreased in MC lines and primary MCTs compared to bone marrow-cultured MCs, suggesting that loss of WWOX is a frequent event in this disease. -

Modeling Enhancer-Promoter Interactions with Attention-Based Neural Networks
bioRxiv preprint doi: https://doi.org/10.1101/219667; this version posted November 14, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Article Modeling Enhancer-Promoter Interactions with Attention-Based Neural Networks Weiguang Mao 1,2, Dennis Kostka 1,2,3 and Maria Chikina 1,2* 1 Department of Computational and Systems Biology, School of Medicine, University of Pittsburgh 2 Joint Carnegie Mellon - University of Pittsburgh Ph.D. Program in Computational Biology 3 Department of Developmental Biology, School of Medicine, University of Pittsburgh * Correspondence: [email protected] Academic Editor: name Version November 9, 2017 submitted to Cells 1 Abstract: Background: Gene regulatory sequences play critical roles in ensuring tightly controlled 2 RNA expression patterns that are essential in a large variety of biological processes. Specifically, 3 enhancer sequences drive expression of their target genes, and the availability of genome-wide 4 maps of enhancer-promoter interactions has opened up the possibility to use machine learning 5 approaches to extract and interpret features that define these interactions in different biological 6 contexts. Methods: Inspired by machine translation models we develop an attention-based neural 7 network model, EPIANN, to predict enhancer-promoter interactions based on DNA sequences. Codes 8 and data are available at https://github.com/wgmao/EPIANN. Results: Our approach accurately 9 predicts enhancer-promoter interactions across six cell lines. In addition, our method generates 10 pairwise attention scores at the sequence level, which specify how short regions in the enhancer and 11 promoter pair-up to drive the interaction prediction.