Table S16:Probe Sets with Significant Variation in Human Datasets from the Met1 Versus DB7 Tumor Comparison Probe Set GSE1456 G
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Meta-Analysis and Inding from Seoul Breast Cancer Study (SEBCS)
The Pharmacogenomics Journal https://doi.org/10.1038/s41397-018-0016-6 ARTICLE Associations between genetic polymorphisms of membrane transporter genes and prognosis after chemotherapy: meta-analysis and finding from Seoul Breast Cancer Study (SEBCS) 1 1 1 1 2 3 4 Ji-Eun Kim ● Jaesung Choi ● JooYong Park ● Chulbum Park ● Se Mi Lee ● Seong Eun Park ● Nan Song ● 5 6 4,7 8 1,4,9 10 Seokang Chung ● Hyuna Sung ● Wonshik Han ● Jong Won Lee ● Sue K. Park ● Mi Kyung Kim ● 4,7 9,11 1,4,9,12 1,4,9 Dong-Young Noh ● Keun-Young Yoo ● Daehee Kang ● Ji-Yeob Choi Received: 7 June 2017 / Revised: 13 October 2017 / Accepted: 4 December 2017 © Macmillan Publishers Limited, part of Springer Nature 2018 Abstract Membrane transporters can be major determinants of the pharmacokinetic profiles of anticancer drugs. The associations between genetic variations of ATP-binding cassette (ABC) and solute carrier (SLC) genes and cancer survival were investigated through a meta-analysis and an association study in the Seoul Breast Cancer Study (SEBCS). Including the SEBCS, the meta-analysis was conducted among 38 studies of genetic variations of transporters on various cancer survivors. 1234567890();,: The population of SEBCS consisted of 1 338 breast cancer patients who had been treated with adjuvant chemotherapy. A total of 7 750 SNPs were selected from 453 ABC and/or SLC genes typed by an Affymetrix 6.0 chip. ABCB1 rs1045642 was associated with poor progression-free survival in a meta-analysis (HR = 1.33, 95% CI: 1.07–1.64). ABCB1, SLC8A1, and SLC12A8 were associated with breast cancer survival in SEBCS (Pgene < 0.05). -

New Products JULY 2013
R&D Systems Tools for Cell Biology Research™ New Products JULY 2013 GMP-grade Recombinant Proteins Contents R&D Systems now offers GMP-grade cytokines and growth factors for research and further manu- facturing applications where current Good Manufacturing Practices (GMP) are required. GMP-grade Recombinant Proteins 2 proteins are manufactured in our ISO-certified facility in compliance with relevant guidelines1 and are produced with extensive documentation at every stage of development from cell culture to final fill and Quantikine® ELISA Kits 3 formulation. GMP-grade Recombinant Human IL-6 and Recombinant Human TNF-a have recently been added to our line of GMP-grade proteins. Additional GMP-grade proteins will be available in the next several months. For an up-to-date product listing or additional information, please visit our website at Luminex® Screening Assays 4 www.RnDSystems.com/GMP. Luminex® Performance Assays 4-5 Features New GMP Proteins ✓ Extensive documentation at every stage of development ProtEin SOURCE Catalog # SIZE Polyclonal Antibodies 6-7 ✓ Documentation of lot-to-lot consistency and traceability Human IL-6 E. coli 206-GMP-010 10 µg Monoclonal Antibodies 7-8 of materials used 206-GMP-050 50 µg ✓ Rigorous quality control using stringent analytical 206-GMP-01M 1 mg processes Biotinylated Antibodies 8 Human TNF-a/ E. coli 210-GMP-010 10 µg ✓ Proven formulations to ensure consistent reconstitution TNFSF1A and results 210-GMP-050 50 µg Antibody Controls 8 210-GMP-01M 1 mg ELISpot Kits & Development Modules 8 30000 60 Fluorokine® Flow Cytometry Kits 8 25000 17348 ) 2 50 20000 Fluorochrome-labeled Antibodies 9 40 8673 DuoSet® ELISA and DuoSet IC ELISA 15000 Development Systems 10 30 Peak Intensity Peak 10000 20 Cell-Based ELISA Assay Kits 10 5000 17565 (Mean RFU x10 Viability Cell 8780 10 Parameter Assay Kits 10 0 0 6000 9000 12000 15000 18000 21000 24000 -3 -2 -1 0 1 10 10 10 10 10 Apoptosis Detection 10 Mass Charge Ratio Recombinant Human TNF-α GMP (ng/mL) MALDI-TOF Analysis of GMP-grade Recombinant Human TNF-a. -

Supplementary Table 1. All Differentially Expressed Genes from Microarray Screening Analysis
Supplementary Table 1. All differentially expressed genes from microarray screening analysis. FCa (Gal-KD Gene Symbol Gene Name p-Value versus Vector) Up-regulated genes HAPLN1 hyaluronan and proteoglycan link protein 1 10.49 0.0027085 THBS1 thrombospondin 1 9.20 0.0022192 ODC1 ornithine decarboxylase 1 6.38 0.0055776 TGM2 transglutaminase 2 4.76 0.0015627 IL7R interleukin 7 receptor 4.75 0.0017245 SERINC2 serine incorporator 2 4.51 0.0014919 ITM2C integral membrane protein 2C 4.32 0.0044644 SERPINB7 serpin peptidase inhibitor, clade B, member 7 4.18 0.0081136 tumor necrosis factor receptor superfamily, member TNFRSF10D 4.01 0.0085561 10d TAGLN transgelin 3.90 0.0099963 LRRN4 leucine rich repeat neuronal 4 3.82 0.0046513 TGFB2 transforming growth factor beta 2 3.51 0.0035017 CPA4 carboxypeptidase A4 3.43 0.0008452 EPB41L3 erythrocyte membrane protein band 4.1-like 3 3.34 0.0025309 NRG1 neuregulin 1 3.28 0.0079724 F3 coagulation factor III (thromboplastin, tissue factor) 3.27 0.0038968 POLR3G polymerase III polypeptide G 3.26 0.0070675 SEMA7A semaphorin 7A 3.20 0.0087335 NT5E 5-nucleotidase 3.17 0.0036353 CAMK2N1 calmodulin-dependent protein kinase II inhibitor 1 3.07 0.0090141 TIMP3 TIMP metallopeptidase inhibitor 3 3.03 0.0047953 SERPINE1 serpin peptidase inhibitor, clade E 2.97 0.0053652 MALL mal, T-cell differentiation protein-like 2.88 0.0078205 DDAH1 dimethylarginine dimethylaminohydrolase 1 2.86 0.0002895 WDR3 WD repeat domain 3 2.85 0.0058842 WNT5A Wnt Family Member 5A 2.81 0.0043796 GPR1 G protein-coupled receptor 1 2.81 0.0021313 -

Supplementary Material Contents
Supplementary Material Contents Immune modulating proteins identified from exosomal samples.....................................................................2 Figure S1: Overlap between exosomal and soluble proteomes.................................................................................... 4 Bacterial strains:..............................................................................................................................................4 Figure S2: Variability between subjects of effects of exosomes on BL21-lux growth.................................................... 5 Figure S3: Early effects of exosomes on growth of BL21 E. coli .................................................................................... 5 Figure S4: Exosomal Lysis............................................................................................................................................ 6 Figure S5: Effect of pH on exosomal action.................................................................................................................. 7 Figure S6: Effect of exosomes on growth of UPEC (pH = 6.5) suspended in exosome-depleted urine supernatant ....... 8 Effective exosomal concentration....................................................................................................................8 Figure S7: Sample constitution for luminometry experiments..................................................................................... 8 Figure S8: Determining effective concentration ......................................................................................................... -

Supplemental Data 5-21-18
Supplemental Methods and Data Androgen receptor polyglutamine expansion drives age-dependent quality control defects and muscle dysfunction Samir R. Nath1,2,3, Zhigang Yu1, Theresa A. Gipson4, Gregory B. Marsh5, Eriko Yoshidome1, Diane M. Robins6, Sokol V. Todi5, David E. Housman4, Andrew P. Lieberman1 1 Department of Pathology, University of Michigan Medical School, Ann Arbor, MI 48109 2 Medical Scientist Training Program, University of Michigan Medical School, Ann Arbor, MI 48109 3 Cellular and Molecular Biology Graduate Program, University of Michigan Medical School, Ann Arbor, MI 48109 4 Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139 5 Department of Pharmacology, Wayne State University School of Medicine, Detroit, MI 48201 6 Department of Human Genetics, University of Michigan Medical School, Ann Arbor, MI 48109 Supplemental Methods qPCR For Drosophila samples, total RNA was extracted from adult fly heads using TRIzol. Fifteen heads were used per sample. Extracted RNA was treated with TURBO DNAse (Ambion) to eliminate contaminating DNA, and reverse transcription was carried out as indicated above. RNA levels were quantified using StepOnePlus Real-Time PCR System with Fast SYBR Green Master Mix (Applied Biosystems). rp49 was used as the internal control. Each round of qRT-PCR was conducted in technical triplicates. A total of three independent repeats was conducted. Fly histology: For histological preparation (75, 76), wings and proboscises of adult flies were removed and bodies were fixed overnight in 2% glutaraldehyde/2% paraformaldehyde in Tris- buffered saline with 0.1% Triton X-100, rotating at 4˚C. Fixed bodies were subsequently dehydrated by using a series of 30%, 50%, 75%, and 100% ethanol/propylene oxide. -

Supporting Information
Supporting Information Table S1. List of confirmed SLC transporters represented in Canine GeneChip. SLC family Members detected Members not detected SLC1: The high affinity glutamate and neutral amino acid SLC1A1 SLC1A2, SLC1A3, SLC1A6 transporter family SLC2: The facilitative GLUT transporter family SLC2A1, SLC2A8 SLC2A3, SLC2A9 SLC3: The heavy subunits of the heteromeric amino acid SLC3A1 transporters SLC4: The bicarbonate transporter family SLC4A11 SLC4A4, SLC4A8 SLC5: The sodium glucose cotransporter family SLC5A6 SLC5A3, SLC5A10, SLC5A12 SLC6: The sodium- and chloride- dependent SLC6A6, SLC6A12 SLCA18 neurotransmitter transporter family SLC7: The cationic amino acid transporter/glycoprotein- NR associated family SLC8: The Na+/Ca2+ exchanger family SLC8A1 SLC9: The Na+/H+ exchanger family SLC9A1, SLC9A6, SLC9A9 SLC10: The sodium bile salt cotransport family SLC10A2 SLC11: The proton coupled metal ion transporter family NR SLC12: The electroneutral cation-Cl cotransporter family SLC12A3, SLC12A6, SLC12A8 SLC13: The human Na+-sulfate/carboxylate cotransporter SLC13A2 family SLC14: The urea transporter family NR SLC15: The proton oligopeptide cotransporter family SLC15A2, SLC15A4 SLC15A1 SLC16: The monocarboxylate transporter family SLC16A13 SLC16A4 SLC17: The vesicular glutamate transporter family SLC17A3, SLC17A7 SLC18: The vesicular amine transporter family NR SLC19: The folate/thiamine transporter family NR SLC20: The type III Na+-phosphate cotransporter family NR SLC21/SLCO: The organic anion transporting family SLC21A3, SLC21A8, -
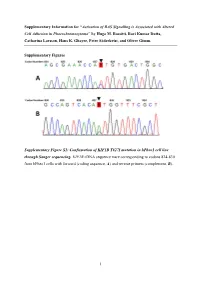
Activation of RAS Signalling Is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M
Supplementary Information for “Activation of RAS Signalling is Associated with Altered Cell Adhesion in Phaeochromocytoma” by Hugo M. Rossitti, Ravi Kumar Dutta, Catharina Larsson, Hans K. Ghayee, Peter Söderkvist, and Oliver Gimm. Supplementary Figures Supplementary Figure S1: Confirmation of KIF1B T827I mutation in hPheo1 cell line through Sanger sequencing. KIF1B cDNA sequence trace corresponding to codons 824-830 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 1 Supplementary Figure S2: Confirmation of NRAS Q61K mutation in hPheo1 cell line through Sanger sequencing. NRAS cDNA sequence trace corresponding to codons 56-67 from hPheo1 cells with forward (coding sequence, A) and reverse primers (complement, B). 2 Supplementary Figure S3: CCND1 gene expression and hPheo1 proliferation. A: Expression of CCND1 mRNA assessed by RT-qPCR and presented as fold change (2-ΔΔCT, mean ± standard error of the mean). B: Cell counts at 1, 2, and 3 days after plating (corresponding to 72, 96 and 120 hours posttransfection, respectively) of control- or siNRAS#1-transfected hPheo1 cells expressed as fold change of the number of cells plated at day 0 (48 hours posttransfection; mean ± standard deviation). All results are from three independent siRNA experiments. 3 Supplementary Tables Supplementary Table S1: List of transcript cluster IDs significantly upregulated in hPheo1 by siNRAS treatment (comparison: siNRAS versus control-transfected hPheo1; ANOVA p < 0.05, FDR < 0.25, fold change < -1.5 or > 1.5). Transcript -

Identification of Placental Nutrient Transporters Associated With
Huang et al. BMC Genomics (2018) 19:173 https://doi.org/10.1186/s12864-018-4518-z RESEARCHARTICLE Open Access Identification of placental nutrient transporters associated with intrauterine growth restriction and pre-eclampsia Xiao Huang1,2, Pascale Anderle3,4, Lu Hostettler2, Marc U. Baumann1,5, Daniel V. Surbek1,5, Edgar C. Ontsouka1,2 and Christiane Albrecht1,2* Abstract Background: Gestational disorders such as intrauterine growth restriction (IUGR) and pre-eclampsia (PE) are main causes of poor perinatal outcomes worldwide. Both diseases are related with impaired materno-fetal nutrient transfer, but the crucial transport mechanisms underlying IUGR and PE are not fully elucidated. In this study, we aimed to identify membrane transporters highly associated with transplacental nutrient deficiencies in IUGR/PE. Results: In silico analyses on the identification of differentially expressed nutrient transporters were conducted using seven eligible microarray datasets (from Gene Expression Omnibus), encompassing control and IUGR/PE placental samples. Thereby 46 out of 434 genes were identified as potentially interesting targets. They are involved in the fetal provision with amino acids, carbohydrates, lipids, vitamins and microelements. Targets of interest were clustered into a substrate-specific interaction network by using Search Tool for the Retrieval of Interacting Genes. The subsequent wet-lab validation was performed using quantitative RT-PCR on placentas from clinically well- characterized IUGR/PE patients (IUGR, n =8;PE,n =5;PE+IUGR,n = 10) and controls (term, n = 13; preterm, n = 7), followed by 2D-hierarchical heatmap generation. Statistical evaluation using Kruskal-Wallis tests was then applied to detect significantly different expression patterns, while scatter plot analysis indicated which transporters were predominantly influenced by IUGR or PE, or equally affected by both diseases. -

RNA-Seq Reveals Conservation of Function Among the Yolk Sacs Of
RNA-seq reveals conservation of function among the PNAS PLUS yolk sacs of human, mouse, and chicken Tereza Cindrova-Daviesa, Eric Jauniauxb, Michael G. Elliota,c, Sungsam Gongd,e, Graham J. Burtona,1, and D. Stephen Charnock-Jonesa,d,e,1,2 aCentre for Trophoblast Research, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, United Kingdom; bElizabeth Garret Anderson Institute for Women’s Health, Faculty of Population Health Sciences, University College London, London, WC1E 6BT, United Kingdom; cSt. John’s College, University of Cambridge, Cambridge, CB2 1TP, United Kingdom; dDepartment of Obstetrics and Gynaecology, University of Cambridge, Cambridge, CB2 0SW, United Kingdom; and eNational Institute for Health Research, Cambridge Comprehensive Biomedical Research Centre, Cambridge, CB2 0QQ, United Kingdom Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved May 5, 2017 (received for review February 14, 2017) The yolk sac is phylogenetically the oldest of the extraembryonic yolk sac plays a critical role during organogenesis (3–5, 8–10), membranes. The human embryo retains a yolk sac, which goes there are limited data to support this claim. Obtaining experi- through primary and secondary phases of development, but its mental data for the human is impossible for ethical reasons, and importance is controversial. Although it is known to synthesize thus we adopted an alternative strategy. Here, we report RNA proteins, its transport functions are widely considered vestigial. sequencing (RNA-seq) data derived from human and murine yolk Here, we report RNA-sequencing (RNA-seq) data for the human sacs and compare them with published data from the yolk sac of and murine yolk sacs and compare those data with data for the the chicken. -

Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared Amongst Δγ T, Innate Lymphoid, and Th Cells
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 δγ is online at: average * The Journal of Immunology , 10 of which you can access for free at: 2016; 197:1460-1470; Prepublished online 6 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600643 http://www.jimmunol.org/content/197/4/1460 Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst T, Innate Lymphoid, and Th Cells You Jeong Lee, Gabriel J. Starrett, Seungeun Thera Lee, Rendong Yang, Christine M. Henzler, Stephen C. Jameson and Kristin A. Hogquist J Immunol cites 41 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2016/07/06/jimmunol.160064 3.DCSupplemental This article http://www.jimmunol.org/content/197/4/1460.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. The Journal of Immunology Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst gd T, Innate Lymphoid, and Th Cells You Jeong Lee,* Gabriel J. -

Transporters
University of Dundee The Concise Guide to PHARMACOLOGY 2015/16 Alexander, Stephen P. H.; Kelly, Eamonn; Marrion, Neil; Peters, John A.; Benson, Helen E.; Faccenda, Elena Published in: British Journal of Pharmacology DOI: 10.1111/bph.13355 Publication date: 2015 Licence: CC BY Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): Alexander, S. P. H., Kelly, E., Marrion, N., Peters, J. A., Benson, H. E., Faccenda, E., Pawson, A. J., Sharman, J. L., Southan, C., Davies, J. A., & CGTP Collaborators (2015). The Concise Guide to PHARMACOLOGY 2015/16: Transporters. British Journal of Pharmacology, 172(24), 6110-6202. https://doi.org/10.1111/bph.13355 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 06. Oct. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: Transporters. -

Wo 2008/156655 A9
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) CORRECTED VERSION (19) World Intellectual Property Organization International Bureau (43) International Publication Date (10) International Publication Number 24 December 2008 (24.12.2008) PCT WO 2008/156655 A9 (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every C07K 14/00 (2006.01) kind of national protection available): AE, AG, AL, AM, AO, AT,AU, AZ, BA, BB, BG, BH, BR, BW, BY, BZ, CA, (21) International Application Number: CH, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, PCT/US2008/007377 EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, (22) International Filing Date: 13 June 2008 (13.06.2008) LK, LR, LS, LT, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PG, PH, (25) Filing Language: English PL, PT, RO, RS, RU, SC, SD, SE, SG, SK, SL, SM, SV, SY, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, (26) Publication Language: English ZA, ZM, ZW (30) Priority Data: (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, 60/934,768 15 June 2007 (15.06.2007) US GM, KE, LS, MW, MZ, NA, SD, SL, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, MD, RU, TJ, TM), (71) Applicants (for all designated States except US): European (AT,BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, GENELUX CORPORATION [US/US]; 3030 Bunker FR, GB, GR, HR, HU, IE, IS, IT, LT,LU, LV,MC, MT, NL, Hill Street, Suite 310, San Diego, CA 92109 (US).