Gram-Positive Anaerobic Cocci: Clinical Relevance, Changed Taxonomy, Identification and Antibiotic Resistance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomics 98 (2011) 370–375
Genomics 98 (2011) 370–375 Contents lists available at ScienceDirect Genomics journal homepage: www.elsevier.com/locate/ygeno Whole-genome comparison clarifies close phylogenetic relationships between the phyla Dictyoglomi and Thermotogae Hiromi Nishida a,⁎, Teruhiko Beppu b, Kenji Ueda b a Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, University of Tokyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan b Life Science Research Center, College of Bioresource Sciences, Nihon University, Fujisawa, Japan article info abstract Article history: The anaerobic thermophilic bacterial genus Dictyoglomus is characterized by the ability to produce useful Received 2 June 2011 enzymes such as amylase, mannanase, and xylanase. Despite the significance, the phylogenetic position of Accepted 1 August 2011 Dictyoglomus has not yet been clarified, since it exhibits ambiguous phylogenetic positions in a single gene Available online 7 August 2011 sequence comparison-based analysis. The number of substitutions at the diverging point of Dictyoglomus is insufficient to show the relationships in a single gene comparison-based analysis. Hence, we studied its Keywords: evolutionary trait based on whole-genome comparison. Both gene content and orthologous protein sequence Whole-genome comparison Dictyoglomus comparisons indicated that Dictyoglomus is most closely related to the phylum Thermotogae and it forms a Bacterial systematics monophyletic group with Coprothermobacter proteolyticus (a constituent of the phylum Firmicutes) and Coprothermobacter proteolyticus Thermotogae. Our findings indicate that C. proteolyticus does not belong to the phylum Firmicutes and that the Thermotogae phylum Dictyoglomi is not closely related to either the phylum Firmicutes or Synergistetes but to the phylum Thermotogae. © 2011 Elsevier Inc. -

Peptoniphilus Stercorisuis Sp. Nov., Isolated from a Swine Manure Storage Tank and Description of Peptoniphilaceae Fam
10848 International Journal of Systematic and Evolutionary Microbiology (2014), 64, 3538–3545 DOI 10.1099/ijs.0.058941-0 Peptoniphilus stercorisuis sp. nov., isolated from a swine manure storage tank and description of Peptoniphilaceae fam. nov. Crystal N. Johnson,1 Terence R. Whitehead,2 Michael A. Cotta,2 Robert E. Rhoades1 and Paul A. Lawson1,3 Correspondence 1Department of Microbiology and Plant Biology, University of Oklahoma, Norman, Paul A. Lawson OK 73019 USA [email protected] 2Bioenergy Research Unit, National Center for Agricultural Utilization Research, USDA, Agricultural Research Service, 1815 N. University Street, Peoria, IL 61604, USA 3Ecology and Evolutionary Biology Program, University of Oklahoma, Norman, OK 73019 USA A species of a previously unknown Gram-positive-staining, anaerobic, coccus-shaped bacterium recovered from a swine manure storage tank was characterized using phenotypic, chemotaxonomic, and molecular taxonomic methods. Comparative 16S rRNA gene sequencing studies and biochemical characteristics demonstrated that this organism is genotypically and phenotypically distinct, and represents a previously unknown sub-line within the order Clostridiales, within the phylum Firmicutes. Pairwise sequence analysis demonstrated that the novel organism clustered within the genus Peptoniphilus, most closely related to Peptoniphilus methioninivorax sharing a 16S rRNA gene sequence similarity of 95.5 %. The major long-chain fatty acids were found to be C14 : 0 (22.4 %), C16 : 0 (15.6 %), C16 : 1v7c (11.3 %) and C16 : 0 ALDE (10.1 %) and the DNA G +C content was 31.8 mol%. Based upon the phenotypic and phylogenetic findings presented, a novel species Peptoniphilus stercorisuis sp. nov. is proposed. The type strain is SF-S1T (5DSM 27563T5NBRC 109839T). -

Metatranscriptomic Analysis Reveals Active Bacterial Communities in Diabetic Foot Infections
fmicb-11-01688 July 20, 2020 Time: 12:22 # 1 ORIGINAL RESEARCH published: 22 July 2020 doi: 10.3389/fmicb.2020.01688 Metatranscriptomic Analysis Reveals Active Bacterial Communities in Diabetic Foot Infections Fatemah Sadeghpour Heravi1, Martha Zakrzewski2, Karen Vickery1, Matthew Malone3,4,5 and Honghua Hu1* 1 Surgical Infection Research Group, Faculty of Medicine and Health Sciences, Macquarie University, Sydney, NSW, Australia, 2 QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia, 3 Infectious Diseases and Microbiology, School of Medicine, Western Sydney University, Sydney, NSW, Australia, 4 Liverpool Hospital, South Western Sydney LHD, Sydney, NSW, Australia, 5 Liverpool Diabetes Collaborative Research Unit, Ingham Institute for Applied Medical Research, Sydney, NSW, Australia Despite the extended view of the composition of diabetic foot infections (DFIs), little is known about which transcriptionally active bacterial communities are pertinent to infection, and if any differences are associated with increased infection severity. We applied a RNA sequencing approach to analyze the composition, function, and pathogenicity of the active bacterial communities in DFIs. Taxonomic profiling of bacterial transcripts revealed the presence of 14 bacterial phyla in DFIs. The abundance Edited by: Qi Zhao, of the Spiroplasma, Vibrio, and Mycoplasma were significantly different in different Liaoning University, China infection severities (P < 0.05). Mild and severe stages of infections were dominated Reviewed by: by Staphylococcus aureus and Porphyromonas asaccharolytica, respectively. A total of Patrick R. M. Harnarayan, The University of the West Indies 132 metabolic pathways were identified of which ribosome and thiamin being among the at St. Augustine, Trinidad and Tobago most highly transcribed pathways. Moreover, a total of 131 antibiotic resistance genes, Maria José Saavedra, primarily involved in the multidrug efflux pumps/exporters, were identified. -

Biofire Blood Culture Identification System (BCID) Fact Sheet
BioFire Blood Culture Identification System (BCID) Fact Sheet What is BioFire BioFire BCID is a multiplex polymerase chain reaction (PCR) test designed to BCID? identify 24 different microorganism targets and three antibiotic resistance genes from positive blood culture bottles. What is the purpose The purpose of BCID is to rapidly identify common microorganisms and of BCID? antibiotic resistance genes from positive blood cultures so that antimicrobial therapy can be quickly optimized by the physician and the antibiotic stewardship pharmacist. It is anticipated that this will result in improved patient outcomes, decreased length of stay, improved antibiotic stewardship, and decreased costs. When will BCID be BCID is performed on all initially positive blood cultures after the gram stain is routinely performed and reported. performed? When will BCID not For blood cultures on the same patient that subsequently become positive with be routinely a microorganism showing the same morphology as the initial positive blood performed? culture, BCID will not be performed. BCID will not be performed on positive blood cultures with gram positive bacilli unless Listeria is suspected. BCID will not be performed on blood culture bottles > 8 hours after becoming positive. BCID will not be performed between 10PM-7AM on weekdays and 2PM-7AM on weekends. BCID will not be performed for clinics that have specifically opted out of testing. How soon will BCID After the blood culture becomes positive and the gram stain is performed and results be available? reported, the bottle will be sent to the core Microbiology lab by routine courier. BCID testing will then be performed. It is anticipated that total turnaround time will generally be 2-3 hours after the gram stain is reported. -

Table 3. Distribution of Tetracycline Resistance Genes Among Gram-Positive Bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces and Ureaplasma Modified Sept
Table 3. Distribution of tetracycline resistance genes among Gram-positive bacteria, Mycobacterium, Mycoplasma, Nocardia, Streptomyces and Ureaplasma Modified Sept. 27, 2021 [n=58 genera] Originally modified from MMBR 2001; 65:232-260 with permission from ASM Journals One Determinant Two Determinants Three or More Determinants n=27 n=7 n=22 k Abiotrophia tet(M) Arthrobacter tet(33)(M) Actinomyces tet(L)(M)(W) Afipia tet(M) Gardnerella tet(M)(Q) Aerococcus tet(M)(O)(58)(61) o Amycolatopsis tet(M) Gemella tet(M)(O) Bacillus tet(K)(L)(M)(O)ao(T)ao(W)(39)m(42)I (45)atotr(A)L Anaerococcus tet(M)g Granulicatella tet(M)(O) Bifidobacterium a, w tet(L)(M)(O)(W) Bacterionema tet(M) Lactococcus tet(M)(S) Bhargavaea tet(L)ac(M)(45)aa ar Brachybacterium tet(M)k Mobiluncusa tet(O)(Q) Clostridiuma,f tet(K)(L)(M)(O)(P)(Q)(W)(36)(40)j(44)p(X) Catenibacteriuma tet(M) Savagea tet(L)(M) Clostridioidesat tet(L)(P)(W)(40) Cellulosimicrobium tet(39)m Corynebacterium tet(M)(Z)(33)(W)q (39)ak Cottaibacterium tet(M) Enterococcus tet(K)(L)(M)(O)(S)(T)(U)(58)ad(61)aq Cutibacterium tet(W)aq Eubacteriuma tet(K)(M)(O)(Q)(32) Erysipelothrix tet(M) Lactobacillusf tet(K)(L)(M)(O)(Q)(S)(W)(Z)(36)am Finegoldia tet(M)g Listeria tet(K)(L)(M)(S)AB(46)ag Geobacillus tet(L) Microbacterium tet(M)(O)ae(42)I Helcococcus tet(M)ah Mycobacteriumc tet(K)(L)(M)(O)t(V)arotr(A)(B) Leifsonia tet(O)t Nocardia tet(K)(L)(M)ai (O) ai Lysinibacillus tet(39)m Paenibacillus tet(L)(M)(O)t(42)i Micrococcus tet(42) Peptostreptococcusa tet(K)(L)(M)(O)(Q) Mycoplasmab tet(M) Sporosarcina tet(K)(L)ac(M)n -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Intraspecies Comparative Genomics of Rickettsia
AIX ͲMARSEILLE UNIVERSITÉ FACULTÉ DE MÉDECINE DE MARSEILLE ÉCOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTÉ T H È S E Présentée et publiquement soutenue devant LA FACULTÉ DE MÉDECINE DE MARSEILLE Le 13 décembre 2013 Par M. Erwin SENTAUSA Né le 16 décembre 1979 àMalang, Indonésie INTRASPECIES COMPARATIVE GENOMICS OF RICKETTSIA Pour obtenir le grade de DOCTORAT d’AIX ͲMARSEILLE UNIVERSITÉ SPÉCIALITÉ :PATHOLOGIE HUMAINE Ͳ MALADIES INFECTIEUSES Membres du Jury de la Thèse : Dr. Patricia RENESTO Rapporteur Pr. Max MAURIN Rapporteur Dr. Florence FENOLLAR Membre du Jury Pr. Pierre ͲEdouard FOURNIER Directeur de thèse Unité de Recherche sur les Maladies Infectieuses et Tropicales Émergentes UM63, CNRS 7278, IRD 198, Inserm 1095 Avant Propos Le format de présentation de cette thèse correspond à une recommandation de la spécialité Maladies Infectieuses et Microbiologie, à l’intérieur du Master de Sciences de la Vie et de la Santé qui dépend de l’Ecole Doctorale des Sciences de la Vie de Marseille. Le candidat est amené àrespecter des règles qui lui sont imposées et qui comportent un format de thèse utilisé dans le Nord de l’Europe permettant un meilleur rangement que les thèses traditionnelles. Par ailleurs, la partie introduction et bibliographie est remplacée par une revue envoyée dans un journal afin de permettre une évaluation extérieure de la qualité de la revue et de permettre àl’étudiant de le commencer le plus tôt possible une bibliographie exhaustive sur le domaine de cette thèse. Par ailleurs, la thèse est présentée sur article publié, accepté ou soumis associé d’un bref commentaire donnant le sens général du travail. -
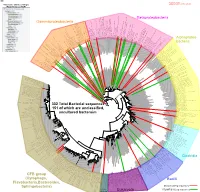
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp. -

Urinicoccus Massiliensis Gen. Nov., Sp. Nov., a New Bacterium Isolated from a Human Urine Sample from a 7-Year-Old Boy Hospitalized for Dental Care
NEW SPECIES Urinicoccus massiliensis gen. nov., sp. nov., a new bacterium isolated from a human urine sample from a 7-year-old boy hospitalized for dental care E. K. Yimagou1, H. Anani2, A. Yacouba1, I. Hasni1, J.-P. Baudoin1, D. Raoult3 and J. Y. Bou Khalil1 1) Aix Marseille University, IRD, AP-HM, MEPHI, 2) Aix Marseille Univ, IRD, AP-HM, VITROME, IHU-Méditerranée Infection, Marseille, France and 3) Special Infectious Agents Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia Abstract Urinicoccus massiliensis strain Marseille-P1992T (= CSURP1992 = DSM100581) is a species of a new genus isolated from human urine. © 2019 The Authors. Published by Elsevier Ltd. Keywords: Culturomics, new species, taxonogenomics, urine, Urinicoccus massiliensis Original Submission: 16 July 2019; Revised Submission: 10 October 2019; Accepted: 17 October 2019 Article published online: 30 October 2019 previously described [7]. The obtained spectra (Fig. 1) were Corresponding author. J. Y. Bou Khalil, MEPHI, Institut Hospitalo- imported into MALDI Biotyper 3.0 software (Bruker Daltonics) Universitaire Méditerranée Infection, 19–21 Boulevard Jean Moulin 13385 Marseille Cedex 05. France and analysed against the main spectra of the bacteria included in E-mail: [email protected] the database (Bruker database constantly updated http://www. mediterraneeinfection.com/article.php?larub=280&titre=urms- database). The initial growth was obtained 10 days after culture on a blood culture vial (Becton Dickinson, Le Pont-de-Claix, Introduction France) supplemented with 5 mL of 0.2-μm-filtered rumen fluid in anaerobic conditions at 37°C and pH 7.5. Culturomics is a concept involving the development of different Strain identification culture conditions in order to enlarge our knowledge of the The 16S rRNA gene was sequenced in order to classify this human microbiota through the discovery of previously uncul- bacterium. -

Identification and Antimicrobial Susceptibility Testing of Anaerobic
antibiotics Review Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Márió Gajdács 1,*, Gabriella Spengler 1 and Edit Urbán 2 1 Department of Medical Microbiology and Immunobiology, Faculty of Medicine, University of Szeged, 6720 Szeged, Hungary; [email protected] 2 Institute of Clinical Microbiology, Faculty of Medicine, University of Szeged, 6725 Szeged, Hungary; [email protected] * Correspondence: [email protected]; Tel.: +36-62-342-843 Academic Editor: Leonard Amaral Received: 28 September 2017; Accepted: 3 November 2017; Published: 7 November 2017 Abstract: Anaerobic bacteria have pivotal roles in the microbiota of humans and they are significant infectious agents involved in many pathological processes, both in immunocompetent and immunocompromised individuals. Their isolation, cultivation and correct identification differs significantly from the workup of aerobic species, although the use of new technologies (e.g., matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, whole genome sequencing) changed anaerobic diagnostics dramatically. In the past, antimicrobial susceptibility of these microorganisms showed predictable patterns and empirical therapy could be safely administered but recently a steady and clear increase in the resistance for several important drugs (β-lactams, clindamycin) has been observed worldwide. For this reason, antimicrobial susceptibility testing of anaerobic isolates for surveillance -

A Genomic Journey Through a Genus of Large DNA Viruses
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Virology Papers Virology, Nebraska Center for 2013 Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses Adrien Jeanniard Aix-Marseille Université David D. Dunigan University of Nebraska-Lincoln, [email protected] James Gurnon University of Nebraska-Lincoln, [email protected] Irina V. Agarkova University of Nebraska-Lincoln, [email protected] Ming Kang University of Nebraska-Lincoln, [email protected] See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/virologypub Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cell and Developmental Biology Commons, Genetics and Genomics Commons, Infectious Disease Commons, Medical Immunology Commons, Medical Pathology Commons, and the Virology Commons Jeanniard, Adrien; Dunigan, David D.; Gurnon, James; Agarkova, Irina V.; Kang, Ming; Vitek, Jason; Duncan, Garry; McClung, O William; Larsen, Megan; Claverie, Jean-Michel; Van Etten, James L.; and Blanc, Guillaume, "Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses" (2013). Virology Papers. 245. https://digitalcommons.unl.edu/virologypub/245 This Article is brought to you for free and open access by the Virology, Nebraska Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Virology Papers by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Adrien Jeanniard, David D. Dunigan, James Gurnon, Irina V. Agarkova, Ming Kang, Jason Vitek, Garry Duncan, O William McClung, Megan Larsen, Jean-Michel Claverie, James L. Van Etten, and Guillaume Blanc This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ virologypub/245 Jeanniard, Dunigan, Gurnon, Agarkova, Kang, Vitek, Duncan, McClung, Larsen, Claverie, Van Etten & Blanc in BMC Genomics (2013) 14. -

(BCID) Results Are “Not Detected”
Interpretation of Positive Blood Cultures When PCR Blood Culture Identification (BCID) Results are “Not Detected” Nebraska Medicine currently uses a multi-plex PCR-based blood culture identification (BCID) system that is able to identify 19 potential pathogens growing in blood culture. BCID generally detects over 90% of the most common causative agents in bloodstream infections; however, when microbes not included on the panel are present in a blood culture, it returns a result of “Not Detected.” This document aims to provide guidance in these scenarios supported by data collected at Nebraska Medicine from January 2018 to August 2019. Table 1: Recommendations for treatment of patients with blood cultures growing organisms not detected on BCID Gram Stain/Preliminary Likely Organism (% total BCID negative)* Recommended Treatment Culture Result Gram-positive: Aerobe Micrococcus sp. (18.1%) (most can also grow in Coagulase-negative Staphylococcus (9.3%) None anaerobic bottles) Diphtheroids (7%) None Peptostreptococcus sp. (4.4%) If therapy is desired: Anaerobe bottle only Lactobacillus sp. (2.6%) Metronidazole 500 mg PO q8h Clostridium sp. (2.6%) OR Penicillin G 4 million units IV q4h Gram-negative: Aerobe Acinetobacter sp. (1.8%) (most can also grow in Stenotrophomonas maltophilia (1.6%) Levofloxacin 750 mg IV/PO q24h anaerobic bottles) Pseudomonas fluorescens-putida group (1%) Bacteroides fragilis group (9.3%) Anaerobe bottle only Metronidazole 500 mg IV/PO q8h Fusobacterium sp. (4.7%) *A full list of isolated organisms can be found below in Table 2 Orange text = Cocci, Blue text = Bacilli (rods) Gram-Positives When BCID results as “Not Detected” but there is microbial growth, the organism is most frequently gram-positive (71%).