(A) Hippocampal Neurons (B) Pre-LCM (C) Post-LCM
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
(12) Patent Application Publication (10) Pub. No.: US 2015/0337275 A1 Pearlman Et Al
US 20150337275A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0337275 A1 Pearlman et al. (43) Pub. Date: Nov. 26, 2015 (54) BOCONVERSION PROCESS FOR Publication Classification PRODUCING NYLON-7, NYLON-7.7 AND POLYESTERS (51) Int. C. CI2N 9/10 (2006.01) (71) Applicant: INVISTATECHNOLOGIES S.a.r.l., CI2P 7/40 (2006.01) St. Gallen (CH) CI2PI3/00 (2006.01) CI2PI3/04 (2006.01) (72) Inventors: Paul S. Pearlman, Thornton, PA (US); CI2P 13/02 (2006.01) Changlin Chen, Cleveland (GB); CI2N 9/16 (2006.01) Adriana L. Botes, Cleveland (GB); Alex CI2N 9/02 (2006.01) Van Eck Conradie, Cleveland (GB); CI2N 9/00 (2006.01) Benjamin D. Herzog, Wichita, KS (US) CI2P 7/44 (2006.01) CI2P I 7/10 (2006.01) (73) Assignee: INVISTATECHNOLOGIES S.a.r.l., (52) U.S. C. St. Gallen (CH) CPC. CI2N 9/13 (2013.01); C12P 7/44 (2013.01); CI2P 7/40 (2013.01); CI2P 13/005 (2013.01); (21) Appl. No.: 14/367,484 CI2P 17/10 (2013.01); CI2P 13/02 (2013.01); CI2N 9/16 (2013.01); CI2N 9/0008 (2013.01); (22) PCT Fled: Dec. 21, 2012 CI2N 9/93 (2013.01); CI2P I3/04 (2013.01); PCT NO.: PCT/US2012/071.472 CI2P 13/001 (2013.01); C12Y 102/0105 (86) (2013.01) S371 (c)(1), (2) Date: Jun. 20, 2014 (57) ABSTRACT Embodiments of the present invention relate to methods for Related U.S. Application Data the biosynthesis of di- or trifunctional C7 alkanes in the (60) Provisional application No. -

Identification of a Novel CHN1 P.(Phe213val) Variant in a Large Han Chinese Family with Congenital Duane Retraction Syndrome
www.nature.com/scientificreports OPEN Identifcation of a novel CHN1 p.(Phe213Val) variant in a large Han Chinese family with congenital Duane retraction syndrome Tai‑Cheng Zhou1,3, Wen‑Hua Duan1,3, Xiao‑Lin Fu2,3, Qin Zhu1, Li‑Yun Guo1, Yuan Zhou1, Zhi‑Juan Hua1, Xue‑Jiao Li1, Dong‑Mei Yang1, Jie‑Ying Zhang1, Jie Yin1, Xiao‑Fan Zhang1, Guang‑Long Zhou1 & Min Hu1* Duane retraction syndrome (DRS) is a neuromuscular dysfunction of the eyes. Although many causative genes of DRS have been identifed in Europe and the United States, few reports have been published in regard to Chinese DRS. The aim of the present study was to explore the genetic defect of DRS in a Chinese family. Exome sequencing was used to identify the disease‑causing gene for the two afected family members. Ophthalmic and physical examinations, as well as genetic screenings for variants in chimerin 1 (CHN1), were performed for all family members. Functional analyses of a CHN1 variant in 293T cells included a Rac‑GTP activation assay, α2‑chimaerin translocation assay, and co‑immunoprecipitation assay. Genetic analysis revealed a NM_001822.7: c.637T > G variant in the CHN1 gene, which resulted in the substitution of a highly conserved C1 domain with valine at codon 213 (NP_001813.1: p.(Phe213Val)) (ClinVar Accession Number: SCV001335305). In-silico analysis revealed that the p.(Phe213Val) substitution afected the protein stability and connections among the amino acids of CHN1 in terms of its tertiary protein structure. Functional studies indicated that the p.(Phe213Val) substitution reduced Rac‑GTP activity and enhanced membrane translocation in response to phorbol‑myristoyl acetate (PMA). -

RT² Profiler PCR Array (384-Well Format) Human Apoptosis 384HT
RT² Profiler PCR Array (384-Well Format) Human Apoptosis 384HT Cat. no. 330231 PAHS-3012ZE For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Applied Biosystems® models 7900HT (384-well block), Format E ViiA™ 7 (384-well block); Bio-Rad CFX384™ RT² Profiler PCR Array, Roche® LightCycler® 480 (384-well block) Format G Description The Human Apoptosis RT² Profiler PCR Array profiles the expression of 370 key genes involved in apoptosis, or programmed cell death. The array includes the TNF ligands and their receptors; members of the bcl-2 family, BIR (baculoviral IAP repeat) domain proteins, CARD domain (caspase recruitment domain) proteins, death domain proteins, TRAF (TNF receptor-associated factor) domain proteins and caspases. These 370 genes are also grouped by their functional contribution to apoptosis, either anti-apoptosis or induction of apoptosis. Using real-time PCR, you can easily and reliably analyze expression of a focused panel of genes related to apoptosis with this array. For further details, consult the RT² Profiler PCR Array Handbook. Sample & Assay Technologies Shipping and storage RT² Profiler PCR Arrays in formats E and G are shipped at ambient temperature, on dry ice, or blue ice packs depending on destination and accompanying products. For long term storage, keep plates at –20°C. Note: Ensure that you have the correct RT² Profiler PCR Array format for your real-time cycler (see table above). Note: Open the package and store the products appropriately immediately -
![Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]](https://docslib.b-cdn.net/cover/8283/computational-genome-wide-identification-of-heat-shock-protein-genes-in-the-bovine-genome-version-1-peer-review-2-approved-1-approved-with-reservations-88283.webp)
Computational Genome-Wide Identification of Heat Shock Protein Genes in the Bovine Genome [Version 1; Peer Review: 2 Approved, 1 Approved with Reservations]
F1000Research 2018, 7:1504 Last updated: 08 AUG 2021 RESEARCH ARTICLE Computational genome-wide identification of heat shock protein genes in the bovine genome [version 1; peer review: 2 approved, 1 approved with reservations] Oyeyemi O. Ajayi1,2, Sunday O. Peters3, Marcos De Donato2,4, Sunday O. Sowande5, Fidalis D.N. Mujibi6, Olanrewaju B. Morenikeji2,7, Bolaji N. Thomas 8, Matthew A. Adeleke 9, Ikhide G. Imumorin2,10,11 1Department of Animal Breeding and Genetics, Federal University of Agriculture, Abeokuta, Nigeria 2International Programs, College of Agriculture and Life Sciences, Cornell University, Ithaca, NY, 14853, USA 3Department of Animal Science, Berry College, Mount Berry, GA, 30149, USA 4Departamento Regional de Bioingenierias, Tecnologico de Monterrey, Escuela de Ingenieria y Ciencias, Queretaro, Mexico 5Department of Animal Production and Health, Federal University of Agriculture, Abeokuta, Nigeria 6Usomi Limited, Nairobi, Kenya 7Department of Animal Production and Health, Federal University of Technology, Akure, Nigeria 8Department of Biomedical Sciences, Rochester Institute of Technology, Rochester, NY, 14623, USA 9School of Life Sciences, University of KwaZulu-Natal, Durban, 4000, South Africa 10School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, 30032, USA 11African Institute of Bioscience Research and Training, Ibadan, Nigeria v1 First published: 20 Sep 2018, 7:1504 Open Peer Review https://doi.org/10.12688/f1000research.16058.1 Latest published: 20 Sep 2018, 7:1504 https://doi.org/10.12688/f1000research.16058.1 Reviewer Status Invited Reviewers Abstract Background: Heat shock proteins (HSPs) are molecular chaperones 1 2 3 known to bind and sequester client proteins under stress. Methods: To identify and better understand some of these proteins, version 1 we carried out a computational genome-wide survey of the bovine 20 Sep 2018 report report report genome. -

Cytokine Nomenclature
RayBiotech, Inc. The protein array pioneer company Cytokine Nomenclature Cytokine Name Official Full Name Genbank Related Names Symbol 4-1BB TNFRSF Tumor necrosis factor NP_001552 CD137, ILA, 4-1BB ligand receptor 9 receptor superfamily .2. member 9 6Ckine CCL21 6-Cysteine Chemokine NM_002989 Small-inducible cytokine A21, Beta chemokine exodus-2, Secondary lymphoid-tissue chemokine, SLC, SCYA21 ACE ACE Angiotensin-converting NP_000780 CD143, DCP, DCP1 enzyme .1. NP_690043 .1. ACE-2 ACE2 Angiotensin-converting NP_068576 ACE-related carboxypeptidase, enzyme 2 .1 Angiotensin-converting enzyme homolog ACTH ACTH Adrenocorticotropic NP_000930 POMC, Pro-opiomelanocortin, hormone .1. Corticotropin-lipotropin, NPP, NP_001030 Melanotropin gamma, Gamma- 333.1 MSH, Potential peptide, Corticotropin, Melanotropin alpha, Alpha-MSH, Corticotropin-like intermediary peptide, CLIP, Lipotropin beta, Beta-LPH, Lipotropin gamma, Gamma-LPH, Melanotropin beta, Beta-MSH, Beta-endorphin, Met-enkephalin ACTHR ACTHR Adrenocorticotropic NP_000520 Melanocortin receptor 2, MC2-R hormone receptor .1 Activin A INHBA Activin A NM_002192 Activin beta-A chain, Erythroid differentiation protein, EDF, INHBA Activin B INHBB Activin B NM_002193 Inhibin beta B chain, Activin beta-B chain Activin C INHBC Activin C NM005538 Inhibin, beta C Activin RIA ACVR1 Activin receptor type-1 NM_001105 Activin receptor type I, ACTR-I, Serine/threonine-protein kinase receptor R1, SKR1, Activin receptor-like kinase 2, ALK-2, TGF-B superfamily receptor type I, TSR-I, ACVRLK2 Activin RIB ACVR1B -

Gene Targeting Therapies (Roy Alcalay)
Recent Developments in Gene - Targeted Therapies for Parkinson’s Disease Roy Alcalay, MD, MS Alfred and Minnie Bressler Associate Professor of Neurology Division of Movement Disorders Columbia University Medical Center Disclosures Funding: Dr. Alcalay is funded by the National Institutes of Health, the DOD, the Michael J. Fox Foundation and the Parkinson’s Foundation. Dr. Alcalay receives consultation fees from Genzyme/Sanofi, Restorbio, Janssen, and Roche. Gene Localizations Identified in PD Gene Symbol Protein Transmission Chromosome PARK1 SNCA α-synuclein AD 4q22.1 PARK2 PRKN parkin (ubiquitin ligase) AR 6q26 PARK3 ? ? AD 2p13 PARK4 SNCA triplication α-synuclein AD 4q22.1 PARK5 UCH-L1 ubiquitin C-terminal AD 4p13 hydrolase-L1 PARK6 PINK1 PTEN-induced kinase 1 AR 1p36.12 PARK7 DJ-1 DJ-1 AR 1p36.23 PARK8 LRRK2 leucine rich repeat kinase 2 AD 12q12 PARK9 ATP13A2 lysosomal ATPase AR 1p36.13 PARK10 ? ? (Iceland) AR 1p32 PARK11 GIGYF2 GRB10-interacting GYF protein 2 AD 2q37.1 PARK12 ? ? X-R Xq21-q25 PARK13 HTRA2 serine protease AD 2p13.1 PARK14 PLA2G6 phospholipase A2 (INAD) AR 22q13.1 PARK15 FBXO7 F-box only protein 7 AR 22q12.3 PARK16 ? Discovered by GWAS ? 1q32 PARK17 VPS35 vacuolar protein sorting 35 AD 16q11.2 PARK18 EIF4G1 initiation of protein synth AD 3q27.1 PARK19 DNAJC6 auxilin AR 1p31.3 PARK20 SYNJ1 synaptojanin 1 AR 21q22.11 PARK21 DNAJC13 8/RME-8 AD 3q22.1 PARK22 CHCHD2 AD 7p11.2 PARK23 VPS13C AR 15q22 Gene Localizations Identified in PD Disorder Symbol Protein Transmission Chromosome PD GBA β-glucocerebrosidase AD 1q21 SCA2 -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -
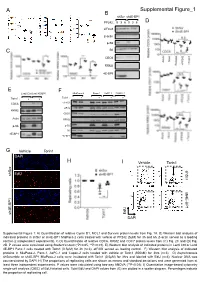
H Supplemental Figure 1 I D E B a C
A Supplemental Figure_1 B shScr sh4E-BP1 PP242 : 0 3 6 0 3 6 D eIF4G1 β-actin p-S6 C S6 CDC6 RRM2 4E-BP1 E F Lenti Ctrl Lenti 4E-BP1 MiaPaca-2 Panc-1 AsPC-1 Capan-2 Torin1 : Torin1 : + + + + + + CDC6 eIF4G1 eIF3a RRM2 CDC6 Actin RRM2 p-S6 p-S6 S6 4E-BP1 4E-BP1 G Vehicle Torin1 DAPI H I Vehicle Torin1 62.20% 45.30% shScr sh4E-BP1 EdU shScr 59.58% 49.47% EdU sh4E-BP1 DAPI Supplemental Figure 1: A) Quantification of relative Cyclin D1, MCL1 and Survivin protein levels from Fig. 1A. B) Western blot analysis of indicated proteins in shScr or sh4E-BP1 MiaPaca-2 cells treated with vehicle or PP242 (5µM) for 3h and 6h. β-actin served as a loading control (2 independent experiments). C-D) Quantification of relative CDC6, RRM2 and CDC7 protein levels from (C) Fig. 2C and (D) Fig. 2D. P values were calculated using Student’s t-test (*P<0.05, **P<0.01). E) Western blot analysis of indicated proteins in Lenti Ctrl or Lenti 4E-BP1 Panc-1 cells treated with Torin1 (0.5µM) for 3h (n=3). eIF4GI served as loading control. F) Western blot analysis of indicated proteins in MiaPaca-2, Panc-1, AsPC-1 and Capan-2 cells treated with vehicle or Torin1 (500nM) for 3hrs (n=3). G) Asynchronous shScramble or sh4E-BP1 MiaPaca-2 cells were incubated with Torin1 (0.5μM) for 3hrs and labeled with EdU (n=3). Nuclear DNA was counterstained by DAPI. H) The proportions of replicating cells are shown as means and standard deviations and were generated from at least three independent experiments. -

Investigation of the Underlying Hub Genes and Molexular Pathogensis in Gastric Cancer by Integrated Bioinformatic Analyses
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Investigation of the underlying hub genes and molexular pathogensis in gastric cancer by integrated bioinformatic analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.20.423656; this version posted December 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The high mortality rate of gastric cancer (GC) is in part due to the absence of initial disclosure of its biomarkers. The recognition of important genes associated in GC is therefore recommended to advance clinical prognosis, diagnosis and and treatment outcomes. The current investigation used the microarray dataset GSE113255 RNA seq data from the Gene Expression Omnibus database to diagnose differentially expressed genes (DEGs). Pathway and gene ontology enrichment analyses were performed, and a proteinprotein interaction network, modules, target genes - miRNA regulatory network and target genes - TF regulatory network were constructed and analyzed. Finally, validation of hub genes was performed. The 1008 DEGs identified consisted of 505 up regulated genes and 503 down regulated genes. -

UNIVERSITY of CALIFORNIA RIVERSIDE Investigations Into The
UNIVERSITY OF CALIFORNIA RIVERSIDE Investigations into the Role of TAF1-mediated Phosphorylation in Gene Regulation A Dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Cell, Molecular and Developmental Biology by Brian James Gadd December 2012 Dissertation Committee: Dr. Xuan Liu, Chairperson Dr. Frank Sauer Dr. Frances M. Sladek Copyright by Brian James Gadd 2012 The Dissertation of Brian James Gadd is approved Committee Chairperson University of California, Riverside Acknowledgments I am thankful to Dr. Liu for her patience and support over the last eight years. I am deeply indebted to my committee members, Dr. Frank Sauer and Dr. Frances Sladek for the insightful comments on my research and this dissertation. Thanks goes out to CMDB, especially Dr. Bachant, Dr. Springer and Kathy Redd for their support. Thanks to all the members of the Liu lab both past and present. A very special thanks to the members of the Sauer lab, including Silvia, Stephane, David, Matt, Stephen, Ninuo, Toby, Josh, Alice, Alex and Flora. You have made all the years here fly by and made them so enjoyable. From the Sladek lab I want to thank Eugene, John, Linh and Karthi. Special thanks go out to all the friends I’ve made over the years here. Chris, Amber, Stephane and David, thank you so much for feeding me, encouraging me and keeping me sane. Thanks to the brothers for all your encouragement and prayers. To any I haven’t mentioned by name, I promise I haven’t forgotten all you’ve done for me during my graduate years. -

Differential Regulation of Estrogen Receptor Α Expression in Breast
Author Manuscript Published OnlineFirst on January 10, 2014; DOI: 10.1158/0008-5472.CAN-13-2020 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. CAN-13-2020R Differential Regulation of Estrogen Receptor α Expression in Breast Cancer Cells by Metastasis-Associated Protein 1 Hyun-Jin Kang1, Min-Ho Lee1, Hae-Lim Kang1, Sung-Hae Kim1, Jung-Ranh Ahn1, Hyelin Na1, Tae-Young Na1, Yona Kim2, Je Kyung Seong2, and Mi-Ock Lee1 1College of Pharmacy and Bio-MAX institute, Research Institute of Pharmaceutical Sciences; and 2College of Veterinary Medicine, BK21 Plus Program for Veterinary Science, Seoul National University, Seoul, Korea; Keywords: Estrogen receptor α; MTA1; IFI16; epigenetics, hormone sensitivity Hyun-Jin Kang, Min-Ho Lee, and Hae-Lim Kang contributed equally to this work. Corresponding Authors: Mi-Ock Lee Ph. D., College of Pharmacy, Seoul National University, 1 Gwanak-ro, Gwanak-gu, Seoul 151-742, Korea. Phone: 82-2-880-9331; Fax: 82-2-887-2692; E-mail: [email protected] Downloaded from cancerres.aacrjournals.org on September 29, 2021. © 2014 American Association for Cancer Research. Author Manuscript Published OnlineFirst on January 10, 2014; DOI: 10.1158/0008-5472.CAN-13-2020 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. ABSTRACT Metastasis-associated protein 1 (MTA1) is a component of the nucleosome remodeling and histone deacetylase (HDAC) complex, which plays an important role in progression of breast cancer. Although MTA1 is known as a repressor of the transactivation function of estrogen receptor (ER)α, its involvement in the epigenetic control of transcription of the ERα gene ESR1 has not been studied.