Promotionsarbeit 2004
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Outline of Presentation Legionella Problem in The
2012/2/9 Legionnaire’s Disease: History, Epidemiology; Assessment and Control of Environmental Outline of Presentation Factors leading to Outbreak Conditions Legionaries’ Disease History Occurrence Nature of Disease Route of Infection Presented at the Pathogenesis HKIOEH Professional Development Seminar Diagnosis and treatment Prevention of infection Understanding the ecology of Legionella February 8, 2012 Control Measures Risk assessment / management Joseph K. Kwan An Outbreak investigation The First Legionnaire’s Disease outbreak It occurred during The 1976 American Legionnaires’ annual meeting at a hotel in Philadelphia, Pennsylvania Upon returning home, 221 got sick, 34 died Investigation revealed a responsible bacteria This Bacterium was named (Legionella pneumophila) Records reveal similar outbreaks 1947 & 1967 It all started here in June 1976 Bellevue-Stratford Hotel in Philadelphia, Pennsylvania, USA Since 1976 Outbreaks continue to Legionella Problem in the USA take place all over the world Between 8000 to 18,000 cases reported each year World-wide Occurrence: 10 to 20 % fatality Range of 1 – 21 cases / million ~ 23% are hospital acquired Europe ~ 4.3 cases / million 30- 40% mortality rate HK ~3 cases / million Accuracy depends on efficiency of recognition and reporting!! ~30,000 patients have died from hospital acquired Legionnaires’ disease in the past 25 years No evidence of transmission from person to person All sources are environment related 1 2012/2/9 The situation in HK Legionella Outbreaks -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Isolation and Identification of Free-Living Amoebae from Tap Water in Sivas, Turkey
Hindawi Publishing Corporation BioMed Research International Volume 2013, Article ID 675145, 8 pages http://dx.doi.org/10.1155/2013/675145 Research Article Isolation and Identification of Free-Living Amoebae from Tap Water in Sivas, Turkey Kübra AçJkalJnCoGkun,1 Semra Özçelik,1 Lütfi Tutar,2 Nazif ElaldJ,3 and Yusuf Tutar4,5 1 Department of Parasitology, Faculty of Medicine, Cumhuriyet University, 58140 Sivas, Turkey 2 Department of Biology, Faculty of Science and Letters, Kahramanmaras¸Sutc¨ ¸u¨ Imam˙ University, 46100 Kahramanmaras, Turkey 3 Department of Infectious Diseases, Faculty of Medicine, Cumhuriyet University, 58140 Sivas, Turkey 4 Department of Biochemistry, Faculty of Pharmacology, Cumhuriyet University, 58140 Sivas, Turkey 5 CUTFAM Research Center, Faculty of Medicine, Cumhuriyet University, 58140 Sivas, Turkey Correspondence should be addressed to Yusuf Tutar; [email protected] Received 9 April 2013; Revised 11 June 2013; Accepted 27 June 2013 Academic Editor: Gernot Zissel Copyright © 2013 Kubra¨ Ac¸ıkalın Cos¸kun et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The present work focuses on a local survey of free-living amoebae (FLA) that cause opportunistic and nonopportunistic infections in humans. Determining the prevalence of FLA in water sources can shine a light on the need to prevent FLA related illnesses. A total of 150 samples of tap water were collected from six districts of Sivas province. The samples were filtered and seeded on nonnutrient agar containing Escherichia coli spread. Thirty-three (22%) out of 150 samples were found to be positive for FLA. -

Subcellular Location of Piscirickettsia Salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis
microorganisms Article Subcellular Location of Piscirickettsia salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis 1, 2,3, 4 5 Cristian Oliver y, Patricio Sánchez y , Karla Valenzuela , Mauricio Hernández , Juan Pablo Pontigo 3, Maria C. Rauch 3, Rafael A. Garduño 4,6 , Ruben Avendaño-Herrera 2,7,* and Alejandro J. Yáñez 2,8,* 1 Laboratorio de Inmunología y Estrés de Organismos Acuáticos, Instituto de Patología Animal, Facultad de Ciencias Veterinarias, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] 2 Interdisciplinary Center for Aquaculture Research, (INCAR), Concepción 4070386, Chile; [email protected] 3 Instituto de Bioquímica y Microbiología, Facultad de Ciencias, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] (J.P.P.); [email protected] (M.C.R.) 4 Microbiology and Immunology Department, Dalhousie University, Halifax, NS B3H 4R2, Canada; [email protected] (K.V.); [email protected] (R.A.G.) 5 Austral-OMICS, Faculty of Sciences, Universidad Austral de Chile, Valdivia 5090000, Chile; [email protected] 6 Canadian Food Inspection Agency, Dartmouth Laboratory, Dartmouth, NS B3B 1Y9, Canada 7 Universidad Andrés Bello, Laboratorio de Patología de Organismos Acuáticos y Biotecnología Acuícola, Facultad Ciencias de la Vida, Viña del Mar 2531015, Chile 8 Facultad de Ciencias, Universidad Austral de Chile, Valdivia 5090000, Chile * Correspondence: [email protected] (R.A.-H.); [email protected] (A.J.Y.) These authors contributed equally to this work. y Received: 12 November 2019; Accepted: 31 December 2019; Published: 15 January 2020 Abstract: Piscirickettsia salmonis is the causative bacterial agent of piscirickettsiosis, a systemic fish disease that significantly impacts the Chilean salmon industry. -

WO 2016/188962 Al 1 December 2016 (01.12.2016) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/188962 Al 1 December 2016 (01.12.2016) P O P C T (51) International Patent Classification: (74) Agents: GOODFELLOW, Hugh Robin et al; Carpmaels C12Q 1/68 (2006.01) & Ransford LLP, One Southampton Row, London WC1B 5HA (GB). (21) International Application Number: PCT/EP2016/061599 (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, (22) Date: International Filing AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, 23 May 20 16 (23.05.2016) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, (25) Filing Language: English DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, (26) Publication Language: English KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (30) Priority Data: MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, 1508860.2 22 May 2015 (22.05.2015) GB PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (71) Applicant: NATIONAL UNIVERSITY OF IRELAND, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. GALWAY [IE/IE]; University Road, Galway (IE). (84) Designated States (unless otherwise indicated, for every (72) Inventors: REDDINGTON, Kate Mary; Deerpack East, kind of regional protection available): ARIPO (BW, GH, Newport Road, Westport, Co. -
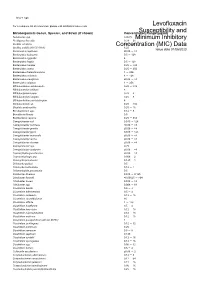
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution. -

HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S
CDAR2L3_IG_HAIRPT_R2_D2_2009FEB HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S. Realm) Draft Standard for Trial Use February 2009 © 2009 Health Level Seven, Inc. Ann Arbor, MI All rights reserved HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 1 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Co-Editor: Marla Albitz Contractor to CDC, Lockheed Martin [email protected] Co-Editor/Co-Chair: Liora Alschuler Alschuler Associates, LLC [email protected] Co-Chair: Calvin Beebe Mayo Clinic [email protected] Co-Chair: Keith W. Boone GE Healthcare [email protected] Co-Editor/Co-Chair: Robert H. Dolin, M.D. Semantically Yours, LLC [email protected] Co-Editor Jonathan Edwards CDC [email protected] Co-Editor Pavla Frazier RN, MSN, MBA Contractor to CDC, Frazier Consulting [email protected] Co-Editor: Sundak Ganesan, M.D. SAIC Consultant to CDC/NCPHI [email protected] Co-Editor: Rick Geimer Alschuler Associates, LLC [email protected] Co-Editor: Gay Giannone M.S.N., R.N. Alschuler Associates LLC [email protected] Co-Editor Austin Kreisler Consultant to CDC/NCPHI [email protected] Primary Editor: Kate Hamilton Alschuler Associates, LLC [email protected] Primary Editor: Brett Marquard Alschuler Associates, LLC [email protected] Co-Editor: Daniel Pollock, M.D. CDC [email protected] HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 2 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Acknowledgments This DSTU was produced and developed in conjunction with the Division of Healthcare Quality Promotion, National Center for Preparedness, Detection, and Control of Infectious Diseases (NCPDCID), and Centers for Disease Control and Prevention (CDC). -

2. Drinking Water Systems in Buildings
g e n liviC tnm apeD apeD r apeD r r nemt nemt t nemt t o t o f o f C f C i C v i v i v l i l i l gnE gnE i gnE i i een een r een r i r i i gn gn gn .sroodni emit rieht fo tsom dneps snamuH snamuH snamuH dneps dneps dneps tsom tsom tsom fo fo fo rieht rieht rieht emit emit emit .sroodni .sroodni .sroodni - o- t o t l aA l aA smetsys st i ht iw tnemnor ivne t l iub er i tne ehT ehT ehT tne i tne i tne er i er er iub l iub t l iub t l t ivne ivne ivne tnemnor tnemnor tnemnor iw iw ht iw ht ht i i st i st st smetsys smetsys smetsys - o t l aA DD DD DD MMicrobiologicalMicrobiologicalicrobiological effectseffectseffects ofofof yaw a ni detarepo dna dengised eb dluohs dluohs dluohs eb eb eb dengised dengised dengised dna dna dna detarepo detarepo detarepo ni ni ni a a a yaw yaw yaw 681 681 681 / / .efas yl lacigoloiborcim si t i taht taht i taht t i t i t si si si lacigoloiborcim lacigoloiborcim yl lacigoloiborcim yl yl .efas .efas .efas / nen inen knI i knI i nneJ i nneJ nen i knI i nneJ 8102 8102 8102 ccoppercopperopper andandand otherotherother abioticabioticabiotic euqinu eht no sesucof siseht sihT sihT sihT siseht siseht siseht sesucof sesucof sesucof no no no eht eht eht euqinu euqinu euqinu n ea nkidf ecnlcgliocim im im lacigoloiborc lacigoloiborc lacigoloiborc sehcin sehcin sehcin fo fo fo gniknird gniknird gniknird retaw retaw retaw dna dna dna gniz i l i tu stnemnor ivne roodni ecaf rus hcuot hcuot hcuot rus rus rus ecaf ecaf ecaf roodni roodni roodni ivne ivne ivne stnemnor stnemnor stnemnor tu i tu l i tu i l i i l i gniz gniz gniz -

Patent (10 ) Patent No
US010195273B2 (12 ) United States Patent (10 ) Patent No. : US 10 , 195 , 273 B2 Clube (45 ) Date of Patent : Feb . 5 , 2019 ( 54 ) SELECTIVELY ALTERING MICROBIOTA 9 , 113 ,616 B2 8 / 2015 MacDonald et al . 9 ,328 , 156 B2 5 /2016 June et al. FOR IMMUNE MODULATION 9 ,464 , 140 B2 10 / 2016 June et al . 9 ,481 , 728 B2 11 / 2016 June et al . (71 ) Applicant : SNIPR TECHNOLOGIES LIMITED , 9 , 499 ,629 B2 11/ 2016 June et al . London (GB ) 9 , 518 , 123 B2 12 / 2016 June et al. 9 , 540 , 445 B2 1 / 2017 June et al . ( 72 ) Inventor: Jasper Clube, London (GB ) 9 , 701, 964 B2 7 / 2017 Clube et al . 2004 /0096974 A1 5 / 2004 Herron et al . 2013 /0109053 Al 5 / 2013 MacDonald et al . (73 ) Assignee : SNIPR TECHNOLOGIES LIMITED , 2013 /0287748 A 10 / 2013 June et al. London (GB ) 2013 /0288368 Al 10 / 2013 June et al. 2013 /0309258 A1 10 / 2013 June et al . ( * ) Notice : Subject to any disclaimer , the term of this 2014 / 0106449 Al 4 / 2014 June et al . patent is extended or adjusted under 35 2014 / 0370017 A1 12 / 2014 June et al. 2015 / 0050699 A1 2 / 2015 Siksnys et al . U . S . C . 154 (b ) by 0 days . 2015 / 0050729 A1 2 / 2015 June et al. 2015 / 0064138 Al 3 / 2015 Lu et al . (21 ) Appl. No. : 15 / 820 ,296 2015 / 0093822 A1 4 / 2015 June et al. 2015 /0099299 Al 4 / 2015 June et al. ( 22 ) Filed : Nov . 21 , 2017 2015 /0118202 A1 4 / 2015 June et al . 2015 /0125463 A1 * 5 /2015 Cogswell . -

Structural and Functional Insights from the Metagenome of an Acidic Hot Spring Microbial Planktonic Community in the Colombian Andes
Structural and Functional Insights from the Metagenome of an Acidic Hot Spring Microbial Planktonic Community in the Colombian Andes Diego Javier Jime´nez1,5*, Fernando Dini Andreote3, Diego Chaves1, Jose´ Salvador Montan˜ a1,2, Cesar Osorio-Forero1,4, Howard Junca1,4, Marı´a Mercedes Zambrano1,4, Sandra Baena1,2 1 Colombian Center for Genomic and Bioinformatics from Extreme Environments (GeBiX), Bogota´, Colombia, 2 Departamento de Biologı´a, Unidad de Saneamiento y Biotecnologı´a Ambiental, Pontificia Universidad Javeriana, Bogota´, Colombia, 3 Department of Soil Science, ‘‘Luiz de Queiroz’’ College of Agriculture, University of Sao Paulo, Piracicaba, Brazil, 4 Molecular Genetics and Microbial Ecology Research Groups, Corporacio´n CorpoGen, Bogota´, Colombia, 5 Department of Microbial Ecology, Center for Ecological and Evolutionary Studies (CEES), University of Groningen, Groningen, The Netherlands Abstract A taxonomic and annotated functional description of microbial life was deduced from 53 Mb of metagenomic sequence retrieved from a planktonic fraction of the Neotropical high Andean (3,973 meters above sea level) acidic hot spring El Coquito (EC). A classification of unassembled metagenomic reads using different databases showed a high proportion of Gammaproteobacteria and Alphaproteobacteria (in total read affiliation), and through taxonomic affiliation of 16S rRNA gene fragments we observed the presence of Proteobacteria, micro-algae chloroplast and Firmicutes. Reads mapped against the genomes Acidiphilium cryptum JF-5, Legionella pneumophila str. Corby and Acidithiobacillus caldus revealed the presence of transposase-like sequences, potentially involved in horizontal gene transfer. Functional annotation and hierarchical comparison with different datasets obtained by pyrosequencing in different ecosystems showed that the microbial community also contained extensive DNA repair systems, possibly to cope with ultraviolet radiation at such high altitudes.