Pathogenic Legionella Spp
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of Biofilms and Protozoa in Legionella Pathogenesis: Implications for Drinking Water
Journal of Applied Microbiology ISSN 1364-5072 REVIEW ARTICLE The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water H.Y. Lau and N.J. Ashbolt National Exposure Research Laboratory, U.S. Environmental Protection Agency, Cincinnati, OH, USA Keywords Summary biofilm, drinking water, protozoa, virulence, water quality. Current models to study Legionella pathogenesis include the use of primary macrophages and monocyte cell lines, various free-living protozoan species and Correspondence murine models of pneumonia. However, there are very few studies of Legionella Helen Y. Lau, U.S. Environmental Protection spp. pathogenesis aimed at associating the role of biofilm colonization and par- Agency, 26 W Martin Luther King Drive, MS asitization of biofilm microbiota and release of virulent bacterial cell ⁄ vacuoles 564, Cincinnati, OH 45268, USA. in drinking water distribution systems. Moreover, the implications of these E-mail: [email protected] environmental niches for drinking water exposure to pathogenic legionellae are 2008 ⁄ 1678: received 30 September 2008, poorly understood. This review summarizes the known mechanisms of Legio- revised 21 November 2008 and accepted 6 nella spp. proliferation within Acanthamoeba and mammalian cells and advo- December 2008 cates the use of the amoeba model to study Legionella pathogenicity because of their close association with Legionella spp. in the aquatic environment. The doi:10.1111/j.1365-2672.2009.04208.x putative role of biofilms and amoebae in the proliferation, development and dissemination of potentially pathogenic Legionella spp. is also discussed. Eluci- dating the mechanisms of Legionella pathogenicity development in our drinking water systems will aid in elimination strategies and procedural designs for drinking water systems and in controlling exposure to Legionella spp. -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -
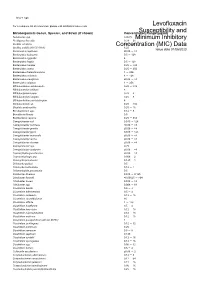
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution. -

Neocallimastix Californiae G1 36,250,970 NA 29,649 95.52 85.2 SRX2598479 (3)
Supplementary material for: Horizontal gene transfer as an indispensable driver for Neocallimastigomycota evolution into a distinct gut-dwelling fungal lineage 1 1 1 2 Chelsea L. Murphy ¶, Noha H. Youssef ¶, Radwa A. Hanafy , MB Couger , Jason E. Stajich3, Y. Wang3, Kristina Baker1, Sumit S. Dagar4, Gareth W. Griffith5, Ibrahim F. Farag1, TM Callaghan6, and Mostafa S. Elshahed1* Table S1. Validation of HGT-identification pipeline using previously published datasets. The frequency of HGT occurrence in the genomes of a filamentous ascomycete and a microsporidian were determined using our pipeline. The results were compared to previously published results. Organism NCBI Assembly Reference Method used Value Value accession number to original in the original reported obtained study study in this study Colletotrichum GCA_000149035.1 (1) Blast and tree 11 11 graminicola building approaches Encephalitozoon GCA_000277815.3 (2) Blast against 12-22 4 hellem custom database, AI score calculation, and tree building Table S2. Results of transcriptomic sequencing. Accession number Genus Species Strain Number of Assembled Predicted peptides % genome Ref. reads transcriptsa (Longest Orfs)b completenessc coveraged (%) Anaeromyces contortus C3G 33,374,692 50,577 22,187 96.55 GGWR00000000 This study Anaeromyces contortus C3J 54,320,879 57,658 26,052 97.24 GGWO00000000 This study Anaeromyces contortus G3G 43,154,980 52,929 21,681 91.38 GGWP00000000 This study Anaeromyces contortus Na 42,857,287 47,378 19,386 93.45 GGWN00000000 This study Anaeromyces contortus O2 60,442,723 62,300 27,322 96.9 GGWQ00000000 This study Anaeromyces robustus S4 21,955,935 NA 17,127 92.41 88.7 SRX3329608 (3) Caecomyces sp. -

HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S
CDAR2L3_IG_HAIRPT_R2_D2_2009FEB HL7 Implementation Guide for CDA Release 2: NHSN Healthcare Associated Infection (HAI) Reports, Release 2 (U.S. Realm) Draft Standard for Trial Use February 2009 © 2009 Health Level Seven, Inc. Ann Arbor, MI All rights reserved HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 1 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Co-Editor: Marla Albitz Contractor to CDC, Lockheed Martin [email protected] Co-Editor/Co-Chair: Liora Alschuler Alschuler Associates, LLC [email protected] Co-Chair: Calvin Beebe Mayo Clinic [email protected] Co-Chair: Keith W. Boone GE Healthcare [email protected] Co-Editor/Co-Chair: Robert H. Dolin, M.D. Semantically Yours, LLC [email protected] Co-Editor Jonathan Edwards CDC [email protected] Co-Editor Pavla Frazier RN, MSN, MBA Contractor to CDC, Frazier Consulting [email protected] Co-Editor: Sundak Ganesan, M.D. SAIC Consultant to CDC/NCPHI [email protected] Co-Editor: Rick Geimer Alschuler Associates, LLC [email protected] Co-Editor: Gay Giannone M.S.N., R.N. Alschuler Associates LLC [email protected] Co-Editor Austin Kreisler Consultant to CDC/NCPHI [email protected] Primary Editor: Kate Hamilton Alschuler Associates, LLC [email protected] Primary Editor: Brett Marquard Alschuler Associates, LLC [email protected] Co-Editor: Daniel Pollock, M.D. CDC [email protected] HL7 Implementation Guide for CDA R2 NHSN HAI Reports Page 2 Draft Standard for Trial Use Release 2 February 2009 © 2009 Health Level Seven, Inc. All rights reserved. Acknowledgments This DSTU was produced and developed in conjunction with the Division of Healthcare Quality Promotion, National Center for Preparedness, Detection, and Control of Infectious Diseases (NCPDCID), and Centers for Disease Control and Prevention (CDC). -

2. Drinking Water Systems in Buildings
g e n liviC tnm apeD apeD r apeD r r nemt nemt t nemt t o t o f o f C f C i C v i v i v l i l i l gnE gnE i gnE i i een een r een r i r i i gn gn gn .sroodni emit rieht fo tsom dneps snamuH snamuH snamuH dneps dneps dneps tsom tsom tsom fo fo fo rieht rieht rieht emit emit emit .sroodni .sroodni .sroodni - o- t o t l aA l aA smetsys st i ht iw tnemnor ivne t l iub er i tne ehT ehT ehT tne i tne i tne er i er er iub l iub t l iub t l t ivne ivne ivne tnemnor tnemnor tnemnor iw iw ht iw ht ht i i st i st st smetsys smetsys smetsys - o t l aA DD DD DD MMicrobiologicalMicrobiologicalicrobiological effectseffectseffects ofofof yaw a ni detarepo dna dengised eb dluohs dluohs dluohs eb eb eb dengised dengised dengised dna dna dna detarepo detarepo detarepo ni ni ni a a a yaw yaw yaw 681 681 681 / / .efas yl lacigoloiborcim si t i taht taht i taht t i t i t si si si lacigoloiborcim lacigoloiborcim yl lacigoloiborcim yl yl .efas .efas .efas / nen inen knI i knI i nneJ i nneJ nen i knI i nneJ 8102 8102 8102 ccoppercopperopper andandand otherotherother abioticabioticabiotic euqinu eht no sesucof siseht sihT sihT sihT siseht siseht siseht sesucof sesucof sesucof no no no eht eht eht euqinu euqinu euqinu n ea nkidf ecnlcgliocim im im lacigoloiborc lacigoloiborc lacigoloiborc sehcin sehcin sehcin fo fo fo gniknird gniknird gniknird retaw retaw retaw dna dna dna gniz i l i tu stnemnor ivne roodni ecaf rus hcuot hcuot hcuot rus rus rus ecaf ecaf ecaf roodni roodni roodni ivne ivne ivne stnemnor stnemnor stnemnor tu i tu l i tu i l i i l i gniz gniz gniz -

Microbial and Mineralogical Characterizations of Soils Collected from the Deep Biosphere of the Former Homestake Gold Mine, South Dakota
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln US Department of Energy Publications U.S. Department of Energy 2010 Microbial and Mineralogical Characterizations of Soils Collected from the Deep Biosphere of the Former Homestake Gold Mine, South Dakota Gurdeep Rastogi South Dakota School of Mines and Technology Shariff Osman Lawrence Berkeley National Laboratory Ravi K. Kukkadapu Pacific Northwest National Laboratory, [email protected] Mark Engelhard Pacific Northwest National Laboratory Parag A. Vaishampayan California Institute of Technology See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/usdoepub Part of the Bioresource and Agricultural Engineering Commons Rastogi, Gurdeep; Osman, Shariff; Kukkadapu, Ravi K.; Engelhard, Mark; Vaishampayan, Parag A.; Andersen, Gary L.; and Sani, Rajesh K., "Microbial and Mineralogical Characterizations of Soils Collected from the Deep Biosphere of the Former Homestake Gold Mine, South Dakota" (2010). US Department of Energy Publications. 170. https://digitalcommons.unl.edu/usdoepub/170 This Article is brought to you for free and open access by the U.S. Department of Energy at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in US Department of Energy Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Gurdeep Rastogi, Shariff Osman, Ravi K. Kukkadapu, Mark Engelhard, Parag A. Vaishampayan, Gary L. Andersen, and Rajesh K. Sani This article is available at DigitalCommons@University of Nebraska - Lincoln: https://digitalcommons.unl.edu/ usdoepub/170 Microb Ecol (2010) 60:539–550 DOI 10.1007/s00248-010-9657-y SOIL MICROBIOLOGY Microbial and Mineralogical Characterizations of Soils Collected from the Deep Biosphere of the Former Homestake Gold Mine, South Dakota Gurdeep Rastogi & Shariff Osman & Ravi Kukkadapu & Mark Engelhard & Parag A. -

Electronic Reporting of Laboratory Data for Public Health
Electronic Reporting of Laboratory Data for Public Health: Meeting Report and Recommendations November 23, 1997 Electronic Reporting of Laboratory Data for Public Health: Meeting Report and Recommendations Report and recommendations endorsed by CDC’s Health Information and Surveillance Systems Board (HISSB) as guidelines for efforts to begin implementation of electronic laboratory reporting. Co-sponsored by: Centers for Disease Control and Prevention Council of State and Territorial Epidemiologists Association of State and Territorial Public Health Laboratory Directors Report prepared by Nicole Lezin and Susan Toal Table of Contents Executive Summary ................................................... i I. Background .........................................................3 Uses of Laboratory and Surveillance Data ...................................3 Efforts to Improve Electronic Laboratory-Based Reporting ......................5 II. Meeting Objectives ...................................................6 III. Goals for Electronic Reporting of Laboratory Data ..........................6 IV. Barriers and Recommended Solutions ....................................8 A. Barriers and Solutions Related to Data Flow ............................8 B. Barriers and Solutions Related to Message Format ......................10 C. Barriers and Solutions Related to Message Content .....................11 V. Next Steps ..........................................................13 Appendix A: Meeting Participants Appendix B: Meeting Agenda Appendix C: Draft -

ESCMID Online Lecture Library © by Author ESCMID Online Lecture Library Latex Agglutination Test
The etiological agent: Legionella pneumophila and other Legionella spp. Valeria Gaia © by author National Reference Centre for Legionella ESCMIDc/o Online Microbiology Lecture laboratory Library Ente Ospedaliero Cantonale Bellinzona - Switzerland © by author ESCMID Online Lecture Library Hystory of Legionnaires’ Disease July 21st 1976 - Philadelphia • 58th Convention of the American Legion at the Bellevue-Stratford Hotel • > 4000 World War II Veterans with families & friends • 600 persons staying at the hotel © by author • ESCMIDJuly 23nd: convention Online closed Lecture Library • Several veterans showed symptoms of pneumonia Searching for the causative agent David Fraser: CDC – Atlanta •Influenza virus? •Nickel intoxication? •Toxin? o 2603 toxicology tests o 5120 microscopy exams o 990 serological tests© by author ESCMIDEverybody seems Online to agree: Lecture it’s NOT a bacterialLibrary disease! July 22nd – August 2nd •High fever •Coughing •Breathing difficulties •Chest pains •Exposed Population =© people by authorstaying in the lobby or outside the Bellevue Stratford Hotel «Broad Street Pneumonia» •221ESCMID persons were Online infected (182+39 Lecture «Broad StreetLibrary Pneumonia» ) 34 patients died (29+5) September 1976-January 1977 Joseph McDade: aims to rule out Q-fever (Rickettsiae) •Injection of “infected” pulmonary tissue in Guinea Pigs microscopy: Cocci and small Bacilli not significant at the time •Inoculation in embryonated eggs + antibiotics to inhibit the growth of contaminating bacteria No growth Microscopy on the -

Legionella and Non-Tuberculous Mycobacteria Using MALDI TOF MS (Matrix Assisted Laser Desorption Ionisation Time of Flight Mass Spectrometry)
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by OTHES DIPLOMARBEIT Titel der Diplomarbeit Establishment of a reference database for Acanthamoeba, Legionella and non-tuberculous mycobacteria using MALDI TOF MS (Matrix Assisted Laser Desorption Ionisation Time of Flight Mass Spectrometry) Verfasserin Dzenita HASANACEVIC angestrebter akademischer Grad Magistra der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. A 442 Studienblatt: Studienrichtung lt. Studienblatt: Anthropologie Betreuerin: Ass. Prof. Univ. Doz. Mag. Dr. Julia Walochnik Contents 1 ABBREVIATIONS ..................................................................................................... 5 2 INTRODUCTION ....................................................................................................... 6 2.1 Acanthamoeba .................................................................................................... 6 2.1.1 Classification ................................................................................................ 6 2.1.1.1 Phylogeny of Acanthamoeba ................................................................. 6 2.1.1.2 Methods of classification ....................................................................... 8 2.1.2 Ecology and geographical distribution ........................................................ 11 2.1.2.1 Life cycle ............................................................................................. 11 2.1.2.2 Trophozoites ......................................................................................