Linkage Map of Escherichia Coli Strain K-12 AUSTIN L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -
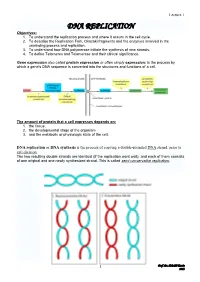
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

An Atpase Domain Common to Prokaryotic Cell Cycle Proteins
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 7290-7294, August 1992 Biochemistry An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp7O heat shock proteins (structural comparison/property pattern/remote homology) PEER BORK, CHRIS SANDER, AND ALFONSO VALENCIA European Molecular Biology Laboratory, D-6900 Heidelberg, Federal Republic of Germany Communicated by Russell F. Doolittle, March 6, 1992 ABSTRACT The functionally diverse actin, hexokinase, and hsp7O protein families have in common an ATPase domain of known three-dimensional structure. Optimal superposition ofthe three structures and alignment ofmany sequences in each of the three families has revealed a set of common conserved residues, distributed in five sequence motifs, which are in- volved in ATP binding and in a putative interdomain hinge. From the multiple sequence aliment in these motifs a pattern of amino acid properties required at each position is defined. The discriminatory power of the pattern is in part due to the use of several known three-dimensional structures and many sequences and in part to the "property" method ofgeneralizing from observed amino acid frequencies to amino acid fitness at each sequence position. A sequence data base search with the pattern significantly matches sugar kinases, such as fuco-, glucono-, xylulo-, ribulo-, and glycerokinase, as well as the prokaryotic cell cycle proteins MreB, FtsA, and StbA. These are predicted to have subdomains with the same tertiary structure as the ATPase subdomains Ia and Ha of hexokinase, actin, and Hsc7O, a very similar ATP binding pocket, and the capacity for interdomain hinge motion accompanying func- tional state changes. -

Non-Homologous Isofunctional Enzymes: a Systematic Analysis Of
Omelchenko et al. Biology Direct 2010, 5:31 http://www.biology-direct.com/content/5/1/31 RESEARCH Open Access Non-homologousResearch isofunctional enzymes: A systematic analysis of alternative solutions in enzyme evolution Marina V Omelchenko, Michael Y Galperin*, Yuri I Wolf and Eugene V Koonin Abstract Background: Evolutionarily unrelated proteins that catalyze the same biochemical reactions are often referred to as analogous - as opposed to homologous - enzymes. The existence of numerous alternative, non-homologous enzyme isoforms presents an interesting evolutionary problem; it also complicates genome-based reconstruction of the metabolic pathways in a variety of organisms. In 1998, a systematic search for analogous enzymes resulted in the identification of 105 Enzyme Commission (EC) numbers that included two or more proteins without detectable sequence similarity to each other, including 34 EC nodes where proteins were known (or predicted) to have distinct structural folds, indicating independent evolutionary origins. In the past 12 years, many putative non-homologous isofunctional enzymes were identified in newly sequenced genomes. In addition, efforts in structural genomics resulted in a vastly improved structural coverage of proteomes, providing for definitive assessment of (non)homologous relationships between proteins. Results: We report the results of a comprehensive search for non-homologous isofunctional enzymes (NISE) that yielded 185 EC nodes with two or more experimentally characterized - or predicted - structurally unrelated proteins. Of these NISE sets, only 74 were from the original 1998 list. Structural assignments of the NISE show over-representation of proteins with the TIM barrel fold and the nucleotide-binding Rossmann fold. From the functional perspective, the set of NISE is enriched in hydrolases, particularly carbohydrate hydrolases, and in enzymes involved in defense against oxidative stress. -

Mutations That Separate the Functions of the Proofreading Subunit of the Escherichia Coli Replicase
G3: Genes|Genomes|Genetics Early Online, published on April 15, 2015 as doi:10.1534/g3.115.017285 Mutations that separate the functions of the proofreading subunit of the Escherichia coli replicase Zakiya Whatley*,1 and Kenneth N Kreuzer*§ *University Program in Genetics & Genomics, Duke University, Durham, NC 27705 §Department of Biochemistry, Duke University Medical Center, Durham, NC 27710 1 © The Author(s) 2013. Published by the Genetics Society of America. Running title: E. coli dnaQ separation of function mutants Keywords: DNA polymerase, epsilon subunit, linker‐scanning mutagenesis, mutation rate, SOS response Corresponding author: Kenneth N Kreuzer, Department of Biochemistry, Box 3711, Nanaline Duke Building, Research Drive, Duke University Medical Center, Durham, NC 27710 Phone: 919 684 6466 FAX: 919 684 6525 Email: [email protected] 1 Present address: Department of Biology, 300 N Washington Street, McCreary Hall, Campus Box 392, Gettysburg College, Gettysburg, PA 17325 Phone: 717 337 6160 Fax: 7171 337 6157 Email: [email protected] 2 ABSTRACT The dnaQ gene of Escherichia coli encodes the ε subunit of DNA polymerase III, which provides the 3’ 5’ exonuclease proofreading activity of the replicative polymerase. Prior studies have shown that loss of ε leads to high mutation frequency, partially constitutive SOS, and poor growth. In addition, a previous study from our lab identified dnaQ knockout mutants in a screen for mutants specifically defective in the SOS response following quinolone (nalidixic acid) treatment. To explain these results, we propose a model whereby in addition to proofreading, ε plays a distinct role in replisome disassembly and/or processing of stalled replication forks. -

WO 2017/014762 Al 26 January 2017 (26.01.2017) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2017/014762 Al 26 January 2017 (26.01.2017) P O P C T (51) International Patent Classification: DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, C12Q 1/68 (2006.01) HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (21) International Application Number: MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PCT/US201 5/0414 15 PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (22) International Filing Date: SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 2 1 July 20 15 (21 .07.2015) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, (71) Applicant: OMNIOME, INC. [US/US]; 4225 Executive TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, Square, Suite 440, La Jolla, California 92037 (US). TJ, TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, (72) Inventors: VIJAYAN, Kandaswamy; 4465 Vision Drive, LV, MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, Unit 6, San Diego, California 92121 (US). -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -

Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals
Journal of the American Heart Association ORIGINAL RESEARCH Association Between the Gut Microbiota and Blood Pressure in a Population Cohort of 6953 Individuals Joonatan Palmu , MD; Aaro Salosensaari , MSc; Aki S. Havulinna , DSc (Tech); Susan Cheng , MD, MPH; Michael Inouye, PhD; Mohit Jain, MD, PhD; Rodolfo A. Salido , BSc; Karenina Sanders , BSc; Caitriona Brennan, BSc; Gregory C. Humphrey, BSc; Jon G. Sanders , PhD; Erkki Vartiainen , MD, PhD; Tiina Laatikainen , MD, PhD; Pekka Jousilahti, MD, PhD; Veikko Salomaa , MD, PhD; Rob Knight , PhD; Leo Lahti , DSc (Tech); Teemu J. Niiranen , MD, PhD BACKGROUND: Several small-scale animal studies have suggested that gut microbiota and blood pressure (BP) are linked. However, results from human studies remain scarce and conflicting. We wanted to elucidate the multivariable-adjusted as- sociation between gut metagenome and BP in a large, representative, well-phenotyped population sample. We performed a focused analysis to examine the previously reported inverse associations between sodium intake and Lactobacillus abun- dance and between Lactobacillus abundance and BP. METHODS AND RESULTS: We studied a population sample of 6953 Finns aged 25 to 74 years (mean age, 49.2±12.9 years; 54.9% women). The participants underwent a health examination, which included BP measurement, stool collection, and 24-hour urine sampling (N=829). Gut microbiota was analyzed using shallow shotgun metagenome sequencing. In age- and sex-adjusted models, the α (within-sample) and β (between-sample) diversities of taxonomic composition were strongly re- lated to BP indexes (P<0.001 for most). In multivariable-adjusted models, β diversity was only associated with diastolic BP (P=0.032). -

A MUTYH Germline Mutation Is Associated with Small Intestinal
248 J P Dumanski et al. MUTYH substitution 24:8 427–443 Research Gly396Asp in SI-NETs Open Access A MUTYH germline mutation is associated with small intestinal neuroendocrine tumors Jan P Dumanski1,*, Chiara Rasi1,*, Peyman Björklund2, Hanna Davies1, Abir S Ali3, Malin Grönberg3, Staffan Welin3, Halfdan Sorbye4,5, Henning Grønbæk6, Janet L Cunningham7, Lars A Forsberg1, Lars Lind8, Erik Ingelsson9, Peter Stålberg10, Per Hellman10 and Eva Tiensuu Janson3 1Department of Immunology, Genetics and Pathology and SciLifeLab, Uppsala University, Uppsala, Sweden 2Department of Surgical Sciences, Experimental Surgery, Uppsala University, Uppsala, Sweden 3Department of Medical Sciences, Endocrine Oncology, Uppsala University, Uppsala, Sweden 4Department of Oncology, Haukeland University Hospital, Bergen, Norway 5Department of Clinical Science, University of Bergen, Bergen, Norway 6Department of Hepatology and Gastroenterology, Aarhus University Hospital, Aarhus, Denmark 7 Department of Neuroscience, Uppsala University, Uppsala, Sweden Correspondence 8 Department of Medical Sciences, Uppsala University, Uppsala, Sweden should be addressed 9 Division of Cardiovascular Medicine, Department of Medicine, Stanford University, San Francisco, California, USA to E Tiensuu Janson 10 Department of Surgical Sciences, Endocrine Surgery, Uppsala University, Uppsala, Sweden Email *(J P Dumanski and C Rasi shared first co-authorship) eva.tiensuu_janson@medsci. uu.se Abstract The genetics behind predisposition to small intestinal neuroendocrine tumors (SI-NETs) Key Words Endocrine-Related Cancer Endocrine-Related is largely unknown, but there is growing awareness of a familial form of the disease. f familial cancer We aimed to identify germline mutations involved in the carcinogenesis of SI-NETs. f cancer predisposition The strategy included next-generation sequencing of exome- and/or whole-genome of f small intestinal carcinoid blood DNA, and in selected cases, tumor DNA, from 24 patients from 15 families with f oxidative stress the history of SI-NETs. -

Supplementary Materials
Supplementary Materials Figure S1. Differentially abundant spots between the mid-log phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 3. Figure S2. Differentially abundant spots between the stationary phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 4. S2 Table S1. Summary of the non-polysaccharide degrading proteins identified in the B. proteoclasticus cytosol by 2DE/MALDI-TOF. Protein Locus Location Score pI kDa Pep. Cov. Amino Acid Biosynthesis Acetylornithine aminotransferase, ArgD Bpr_I1809 C 1.7 × 10−4 5.1 43.9 11 34% Aspartate/tyrosine/aromatic aminotransferase Bpr_I2631 C 3.0 × 10−14 4.7 43.8 15 46% Aspartate-semialdehyde dehydrogenase, Asd Bpr_I1664 C 7.6 × 10−18 5.5 40.1 17 50% Branched-chain amino acid aminotransferase, IlvE Bpr_I1650 C 2.4 × 10−12 5.2 39.2 13 32% Cysteine synthase, CysK Bpr_I1089 C 1.9 × 10−13 5.0 32.3 18 72% Diaminopimelate dehydrogenase Bpr_I0298 C 9.6 × 10−16 5.6 35.8 16 49% Dihydrodipicolinate reductase, DapB Bpr_I2453 C 2.7 × 10−6 4.9 27.0 9 46% Glu/Leu/Phe/Val dehydrogenase Bpr_I2129 C 1.2 × 10−30 5.4 48.6 31 64% Imidazole glycerol phosphate synthase Bpr_I1240 C 8.0 × 10−3 4.7 22.5 8 44% glutamine amidotransferase subunit Ketol-acid reductoisomerase, IlvC Bpr_I1657 C 3.8 × 10−16 -

Microbial (Per)Chlorate Reduction in Hot Subsurface Environments
Microbial (Per)chlorate Reduction in Hot Subsurface Environments Martin G. Liebensteiner Thesis committee Promotor Prof. Dr Alfons J.M. Stams Personal chair at the Laboratory of Microbiology Wageningen University Co-promotor Dr Bart P. Lomans Principal Scientist Shell Global Solutions International B.V., Rijswijk Other members Prof. Dr Willem J.H. van Berkel, Wageningen University Prof. Dr Mike S.M. Jetten, Radboud University Nijmegen Prof. Dr Ian Head, Newcastle University, UK Dr Timo J. Heimovaara, Delft University of Technology This research was conducted under the auspices of the Graduate School for Socio-Economic and Natural Sciences of the Environment (SENSE) Microbial (Per)chlorate Reduction in Hot Subsurface Environments Martin G. Liebensteiner Thesis submitted in fulfi lment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnifi cus Prof. Dr M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 17 October 2014 at 4 p.m. in the Aula. Martin G. Liebensteiner Microbial (Per)chlorate Reduction in Hot Subsurface Environments 172 pages. PhD thesis, Wageningen University, Wageningen, NL (2014) With references, with summaries in Dutch and English ISBN 978-94-6257-125-9 TABLE OF CONTENTS Chapter 1 General introduction and thesis outline 7 Chapter 2 Microbial redox processes in deep subsurface environments 23 and the potential application of (per)chlorate in oil reservoirs Chapter 3 (Per)chlorate reduction by the hyperthermophilic