Systematics of the Ameerega Rubriventris Complex (Anura: Dendrobatidae) with Descriptions of Two New Cryptic Species from the East-Andean Versant of Peru
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Global Models of Ant Diversity Suggest Regions Where New Discoveries Are Most Likely Are Under Disproportionate Deforestation Threat
Global models of ant diversity suggest regions where new discoveries are most likely are under disproportionate deforestation threat Benoit Guénard1, Michael D. Weiser, and Robert R. Dunn Department of Biology and the Keck Center for Behavioral Biology, North Carolina State University, Raleigh, NC 27695 Edited* by Edward O. Wilson, Harvard University, Cambridge, MA, and approved March 23, 2012 (received for review August 24, 2011) Most of the described and probably undescribed species on Earth As for other taxa, richness decreased with latitude (e.g., refs. 13 are insects. Global models of species diversity rarely focus on and 14) (Fig. 1) and there were also strong regional effects on the insects and none attempt to address unknown, undescribed magnitude of diversity. African regions were less diverse than diversity. We assembled a database representing about 13,000 would be expected given their latitude (or climate), the reverse of records for ant generic distribution from over 350 regions that the pattern observed for termites (15, 16) and terrestrial mammals cover much of the globe. Based on two models of diversity and (17), although similar to that for vascular plants (5, 18). The 53 endemicity, we identified regions where our knowledge of ant endemic genera were found almost exclusively in tropical regions diversity is most limited, regions we have called “hotspots of dis- that were diverse more generally, with four interesting exceptions covery.” A priori, such regions might be expected to be remote in North Africa, Armenia, Azerbaijan, and South Korea (Fig. 2). and untouched. Instead, we found that the hotspots of discovery Both overall generic diversity and endemic diversity showed are also the regions in which biodiversity is the most threatened a peak in the Oriental region, especially in Borneo, and were also by habitat destruction. -

DNA Barcoding the Smaller Arachnid Orders from ACP Panguana, Amazonian Peru
SPIXIANA 41 2 169-172 München, Dezember 2018 ISSN 0341-8391 DNA barcoding the smaller arachnid orders from ACP Panguana, Amazonian Peru (Amblypygi, Phrynidae and Schizomida, Hubbardiidae) Tobias Lehmann & Stefan Friedrich Lehmann, T. & Friedrich, S. 2018. DNA barcoding the smaller arachnid orders from ACP Panguana, Amazonian Peru (Amblypygi, Phrynidae and Schizomida, Hubbardiidae). Spixiana 41 (2): 169-172. Amblypygi and Schizomida were collected at the private protected area ACP Panguana, located in the primary evergreen lowland rainforest of Amazonian Peru. Through integrative taxonomy, using COI barcoding and morphological determi- nation, all amblypygids could be identified as Heterophrynus elaphus Pocock, 1903, and the schizomids as Surazomus chavin Pinto-da-Rocha, 1996. COI p-distances are provided and DNA sequences were uploaded to BOLD. Tobias Lehmann (corresponding author), SNSB – ZSM, Bavarian State Collection of Zoology, Münchhausenstr. 21, 81247 Munich, Germany; and Ludwig-Maximili- ans-Universität München, Department Biologie II, Großhaderner Str. 2, 82152 Planegg-Martinsried, Germany; e-mail: [email protected] Stefan Friedrich, SNSB – ZSM, Bavarian State Collection of Zoology, Münch- hausenstr. 21, 81247 Munich, Germany Introduction beds and on large trees. Here, two different colour morphs were found: one large generally brownish Amblypygi, Thelyphonida, Schizomida, Palpigradi, one and one smaller one with a whitish front part Ricinulei and Solifugae are often combined as the of the prosoma and reddish pedipalps. The smaller “Smaller Arachnid Orders” (Harvey 2003, Harvey specimens always showed the white and red col- 2007). Recently an inventory of the soil arthropod ouration and the larger specimens were generally fauna of the biological field station and private pro- brown, no intermediate morphs were found. -

Chromosome Analysis of Five Brazilian
c Indian Academy of Sciences RESEARCH ARTICLE Chromosome analysis of five Brazilian species of poison frogs (Anura: Dendrobatidae) PAULA CAMARGO RODRIGUES1, ODAIR AGUIAR2, FLÁVIA SERPIERI1, ALBERTINA PIMENTEL LIMA3, MASAO UETANEBARO4 and SHIRLEI MARIA RECCO-PIMENTEL1∗ 1Departamento de Anatomia, Biologia Celular e Fisiologia, Instituto de Biologia, Universidade Estadual de Campinas, 13083-863 Campinas, São Paulo, Brazil 2Departamento de Biociências, Universidade Federal de São Paulo, Campus Baixada Santista, 11060-001 Santos, São Paulo, Brazil 3Coordenadoria de Pesquisas em Ecologia, Instituto Nacional de Pesquisas do Amazonas, 69011-970 Manaus, Amazonas, Brazil 4Departamento de Biologia, Universidade Federal de Mato Grosso do Sul, 70070-900 Campo Grande, Mato Grosso do Sul, Brazil Abstract Dendrobatid frogs have undergone an extensive systematic reorganization based on recent molecular findings. The present work describes karyotypes of the Brazilian species Adelphobates castaneoticus, A. quinquevittatus, Ameerega picta, A. galactonotus and Dendrobates tinctorius which were compared to each other and with previously described related species. All karyotypes consisted of 2n = 18 chromosomes, except for A. picta which had 2n = 24. The karyotypes of the Adelphobates and D. tinctorius species were highly similar to each other and to the other 2n = 18 previously studied species, revealing conserved karyotypic characteristics in both genera. In recent phylogenetic studies, all Adelphobates species were grouped in a clade separated from the Dendrobates species. Thus, we hypothesized that their common karyotypic traits may have a distinct origin by chromosome rearrangements and mutations. In A. picta, with 2n = 24, chromosome features of pairs from 1 to 8 are shared with other previously karyotyped species within this genus. Hence, the A. -
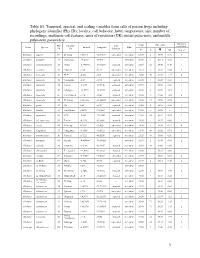
1 Table S1. Temporal, Spectral, and Scaling Variables from Calls Of
Table S1. Temporal, spectral, and scaling variables from calls of poison frogs including phylogeny identifier (Phy ID), locality, call behavior, habit, temperature, size, number of recordings, multinote call features, units of repetition (UR), initial pulse-note, and middle pulse-note parameters. Analyzed Phy Locality Call Temp SVL (mm) Genus Species Latitude Longitude Habit recordings ID ID Behavior °C N ! SD N of ♂ Allobates algorei 60 El Tama 7.65375 -72.19137 concealed terrestrial 23.50 8 18.90 0.70 3 Allobates brunneus 37 Guimaraes -15.2667 -55.5311 -- terrestrial 26.50 1 16.13 0.00 1 Allobates caeruleodactylus 48 Borba -4.398593 -59.60251 exposed terrestrial 25.60 12 15.50 0.40 1 Allobates crombiei 52 Altamira -3.65 -52.38 concealed terrestrial 24.10 2 18.10 0.04 2 Allobates femoralis 43 ECY -0.633 -76.5 concealed terrestrial 25.60 20 23.58 1.27 6 Allobates femoralis 46 Porongaba -8.67 -72.78 exposed terrestrial 25.00 1 25.38 0.00 1 Allobates femoralis 44 Leticia -4.2153 -69.9406 exposed terrestrial 25.50 1 20.90 0.00 1 Allobates femoralis 40 Albergue -12.8773 -71.3865 exposed terrestrial 26.00 6 21.98 2.18 1 Allobates femoralis 41 CAmazonico -12.6 -70.08 exposed terrestrial 26.00 12 22.43 1.06 4 Allobates femoralis 45 El Palmar 8.333333 -61.66667 concealed terrestrial 24.00 27 25.50 0.76 1 Allobates granti 49 FG 3.62 -53.17 exposed terrestrial 24.60 8 16.15 0.55 1 Allobates humilis 59 San Ramon 8.8678 -70.4861 concealed terrestrial 19.50 -- 21.80 -- 1 Allobates insperatus 54 ECY -0.633 -76.4005 exposed terrestrial 24.60 18 16.64 0.93 7 Allobates aff. -

Taxonomic Checklist of Amphibian Species Listed in the CITES
CoP17 Doc. 81.1 Annex 5 (English only / Únicamente en inglés / Seulement en anglais) Taxonomic Checklist of Amphibian Species listed in the CITES Appendices and the Annexes of EC Regulation 338/97 Species information extracted from FROST, D. R. (2015) "Amphibian Species of the World, an online Reference" V. 6.0 (as of May 2015) Copyright © 1998-2015, Darrel Frost and TheAmericanMuseum of Natural History. All Rights Reserved. Additional comments included by the Nomenclature Specialist of the CITES Animals Committee (indicated by "NC comment") Reproduction for commercial purposes prohibited. CoP17 Doc. 81.1 Annex 5 - p. 1 Amphibian Species covered by this Checklist listed by listed by CITES EC- as well as Family Species Regulation EC 338/97 Regulation only 338/97 ANURA Aromobatidae Allobates femoralis X Aromobatidae Allobates hodli X Aromobatidae Allobates myersi X Aromobatidae Allobates zaparo X Aromobatidae Anomaloglossus rufulus X Bufonidae Altiphrynoides malcolmi X Bufonidae Altiphrynoides osgoodi X Bufonidae Amietophrynus channingi X Bufonidae Amietophrynus superciliaris X Bufonidae Atelopus zeteki X Bufonidae Incilius periglenes X Bufonidae Nectophrynoides asperginis X Bufonidae Nectophrynoides cryptus X Bufonidae Nectophrynoides frontierei X Bufonidae Nectophrynoides laevis X Bufonidae Nectophrynoides laticeps X Bufonidae Nectophrynoides minutus X Bufonidae Nectophrynoides paulae X Bufonidae Nectophrynoides poyntoni X Bufonidae Nectophrynoides pseudotornieri X Bufonidae Nectophrynoides tornieri X Bufonidae Nectophrynoides vestergaardi -

Reproductive Biology of Ameerega Trivittata(Anura: Dendrobatidae)
ACTA AMAZONICA http://dx.doi.org/10.1590/1809-4392201305384 Reproductive biology of Ameerega trivittata (Anura: Dendrobatidae) in an area of terra firme forest in eastern Amazonia Ellen Cristina Serrão ACIOLI1 * , Selvino NECKEL-OLIVEIRA2 ¹ Universidade Federal do Pará/Museu Paraense Emílio Goeldi, Programa de Pós Graduação em Zoologia, Av. Perimetral 1901/1907, Terra Firme, 66017-970, Belém, Pará, Brasil. ² Universidade Federal de Santa Catarina, Departamento de Ecologia e Zoologia, Centro de Ciências Biológicas, 88040-970, Florianópolis, Santa Catarina, Brasil. * Corresponding author: [email protected] ABSTRACT The reproductive success of tropical amphibians is influenced by factors such as body size and the characteristics of breeding sites. Data on reproductive biology are important for the understanding of population dynamics and the maintenance of species. The objectives of the present study were to examine the abundance ofAmeerega trivittata, analyze the use of microhabitats by calling males and the snout-vent length (SVL) of breeding males and females, the number of tadpoles carried by the males and mature oocytes in the females, as well as the relationship between the SVL of the female and both the number and mean size of the mature oocytes found in the ovaries. Three field trips were conducted between January and September, 2009. A total of 31 plots, with a mean area of 2.3 ha, were surveyed, resulting in records of 235 individuals, with a mean density of 3.26 individuals per hectare. Overall, 66.1% of the individuals sighted were located in the leaf litter, while 17.4% were perched on decaying tree trunks on the forest floor, 15.7% on the aerial roots ofCecropia trees, and 0.8% on lianas. -

Diet Composition of Ameerega Picta
Bonn zoological Bulletin 68 (1): 93–96 ISSN 2190–7307 2019 · Landgref Filho P. et al. http://www.zoologicalbulletin.de https://doi.org/10.20363/BZB-2019.68.1.093 Scientific note urn:lsid:zoobank.org:pub:CBA156F4-E9AA-401C-B036-77C362CE1E89 Diet composition of Ameerega picta (Tschudi, 1838) from the Serra da Bodoquena region in central Brazil, with a summary of dietary studies on species of the genus Ameerega (Anura: Dendrobatidae) Paulo Landgref Filho1, Fabrício H. Oda2, *, Fabio T. Mise3, Domingos de J. Rodrigues4 & Masao Uetanabaro5 1 Universidade Federal de Mato Grosso do Sul, Campus Aquidauana, 79200-000, Aquidauana, Mato Grosso do Sul, Brazil 2 Departamento de Química Biológica, Programa de Pós-graduação em Bioprospecção Molecular, Universidade Regional do Cariri, 63105-00, Crato, Ceará, Brazil 3 Departamento de Biologia, Universidade Estadual do Centro-Oeste, 85040-080, Guarapuava, Paraná, Brazil 4 Instituto de Ciências Naturais, Humanas e Sociais, Universidade Federal de Mato Grosso – Campus Universitário de Sinop, 78557-267, Sinop, Mato Grosso, Brazil 4 Instituto Nacional de Ciência e Tecnologia de Estudos Integrados da Biodiversidade Amazônica – Núcleo Regional de Sinop, Sinop, Mato Grosso, Brazil 5 Rua Clóvis n. 24, 79022-071, Campo Grande, Mato Grosso do Sul, Brazil * Corresponding author: Email: [email protected] 1 urn:lsid:zoobank.org:author:5821C96D-4D9E-4E1D-BE9B-C231B77D1AF3 2 urn:lsid:zoobank.org:author:349712B5-E1B2-4059-A826-58F081DD3A4D 3 urn:lsid:zoobank.org:author:E996AF2A-6CB7-4B73-8DEF-AB7C5EEE1AA0 4 urn:lsid:zoobank.org:author:ACFBA432-3220-4910-AFF5-8FF85053A872 5 urn:lsid:zoobank.org:author:626F6AE3-7D90-4500-BFDC-4E6F4E01B378 Abstract. -

Deforestation by Definition
DEFORESTATION BY DEFINITION THE PERUVIAN GOVERNMENT FAILS TO DEFINE FORESTS AS FORESTS, WHILE PALM OIL EXPANSION AND THE MALAYSIAN INFLUENCE THREATEN THE AMAZON CONTENTS 3 EXECUTIVE SUMMARY 5 1. GRUPO ROMERO: PLANNED DEFORESTATION 8 1.1 HOW A SKEWED FOREST DEFINITION RESULTS IN DEFORESTATION 12 1.2 VIOLATION OF RESERVE REQUIREMENTS EIA would like to thank the following organizations and 13 1.3 AN INVALID LAND TRANSFER individuals for contributions to this report: 15 1.4 GRUPO ROMERO EXISTING PALM OIL PLANTATIONS Asociación Interétnica de la Selva Peruana (AIDESEP) 20 2. MELKA GROUP: AMASSING LAND IN THE AMAZON Andrew Heatherington 24 2.1 LOOMING DEFORESTATION: 458 PROPERTIES AND COUNTING Bruno Manser Fund 26 2.2 MELKA GROUP’S ONGOING DEFORESTATION: TAMSHIYACU AND NUEVA REQUENA Center for International Environmental Law 32 2.3 ILLEGALITIES IN TAMSHIYACU AND NUEVA REQUENA Clinton Jenkins 35 2.4 INSTITUTIONAL LIMITATIONS: THE GOVERNMENT’S INABILITY TO STOP DEFORESTATION FOR MONOCULTURE PLANTATIONS Global Witness Juan Luis Dammert 38 3. GREASING THE PALMS: DENNIS MELKA, ASIAN PLANTATIONS LTD., AND FOREST DESTRUCTION IN SARAWAK, MALAYSIA Nick Cuba 40 3.1 A NEW EMPIRE OF DEFORESTATION Oxfam 41 3.2 ASIAN PLANTATIONS LTD. Sam Lawson 47 3.3 KERESA PLANTATIONS: GRAEME BROWN, THE LINGGI FAMILY, AND Sidney Novoa CLEARCUTTING FOR OIL PALM Transparent World 52 3.4 RSPO-CERTIFIED FOREST DESTRUCTION Henry Túpac Espíritu 53 3.5 ASIAN PLANTATIONS LTD’S SUBSIDIARIES IN SARAWAK: VARIATIONS ON A THEME The local residents of Barranquita, Nueva Requena, 57 3.6 ASIAN PLANTATIONS LTD.’S MODEL OF INTERNATIONAL FINANCING FOR OIL PALM Shanusi and Tamshiyacu 62 CONCLUSION EIA would also like the thank the following funders for their support: 64 RECOMMENDATIONS Cox Foundation 67 GLOSSARY OF TERMS AND ACRONYMS Good Energies Foundation 72 MAPPING DEFORESTATION: ONGOING AND PROJECTED Lia Foundation 74 ANNEXES Overbrook Foundation Tilia Foundation 84 WORKS CITED Weeden Foundation BOXES EIA is responsible for the content of this report ©Environmental Investigation Agency 2015. -

Twenty-Fifth Meeting of the Animals Committee
AC25 Doc. 22 (Rev. 1) Annex 3 (English only / únicamente en inglés / seulement en anglais) Annex 3 Fauna: new species and other changes relating to species listed in the EC wildlife trade regulations – Report compiled by UNEP-WCMC to the European Commission, March, 2011 AC25 Doc. 22 (Rev. 1) Annex 3 – p. 1 Fauna: new species and other taxonomic changes relating to species listed in the EC wildlife trade regulations March, 2011 A report to the European Commission Directorate General E - Environment ENV.E.2. – Environmental Agreements and Trade by the United Nations Environment Programme World Conservation Monitoring Centre AC25 Doc. 22 (Rev. 1) Annex 3 – p. 2 UNEP World Conservation Monitoring Centre 219 Huntingdon Road Cambridge CB3 0DL United Kingdom Tel: +44 (0) 1223 277314 Fax: +44 (0) 1223 277136 Email: [email protected] Website: www.unep-wcmc.org CITATION UNEP-WCMC. 2011. Fauna: new species and other taxonomic changes relating to species ABOUT UNEP-WORLD CONSERVATION listed in the EC wildlife trade regulations. A MONITORING CENTRE report to the European Commission. UNEP- The UNEP World Conservation Monitoring WCMC, Cambridge. Centre (UNEP-WCMC), based in Cambridge, UK, is the specialist biodiversity information and assessment centre of the United Nations Environment Programme (UNEP), run PREPARED FOR cooperatively with WCMC, a UK charity. The The European Commission, Brussels, Belgium Centre's mission is to evaluate and highlight the many values of biodiversity and put authoritative biodiversity knowledge at the DISCLAIMER centre of decision-making. Through the analysis and synthesis of global biodiversity knowledge The contents of this report do not necessarily the Centre provides authoritative, strategic and reflect the views or policies of UNEP or timely information for conventions, countries contributory organisations. -

Usaid/Peru 118/119 Tropical Forest and Biodiversity Analysis
DIEGO PÉREZ USAID/PERU 118/119 TROPICAL FOREST AND BIODIVERSITY ANALYSIS Report authors: Juan Carlos Riveros, Maina Martir-Torres, César Ipenza, Patricia Tello September, 2019 DISCLAIMER: The author’s views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States Government. USAID/PERU 118/119 TROPICAL FOREST AND BIODIVERSITY ANALYSIS September, 2019 Prepared with technical support from US Forest Service International Programs LIST OF FIGURES LIST OF MAPS Figure 1 Map 1 Summary of Main Threats and Drivers of Official Ecosystems Map of Peru 32 Biodiversity and Tropical Forest Loss in Tropical Forests and Marine Ecosystems 13 Map 2 Forest Loss in the Peruvian Amazon Figure 2 Between 2001-2017 39 Forest Loss in Peru 38 Map 3 Figure 3 National Natural Protected Areas Species Richness of Select Taxonomic Managed by SERNANP 43 Groups in Peru 40 Map 4 Figure 4 Forest Use Designations 45 Number of Threatened Plant Species 41 Figure 5 Number of Threatened Animal Species 41 Figure A5 1 Forest Loss in Selected Regions 135 LIST OF TABLES Table 1 Table A2 1 Actions Necessary to Conserve Biodiversity Weekly Activities and Milestones 118 (Tropical Forests and Marine Ecosystems) 15 Table A5 1 Table 2 Ecosystem Categories 128 Policies and Other Legal Instruments Relevant for Biodiversity and Tropical Table A5 2 Forest Conservation 59 National Natural Protected Areas 130 Table 3 Table A5 3 Actions Necessary to Conserve Biodiversity CITES Listed Animal Species 133 (Tropical -

ANFIBIOS De Cordillera Azul
Parque Nacional Cordillera Azul, San Martín, Loreto, Ucayali y Huánuco, PERÚ 1 ANFIBIOS de Cordillera Azul Kaitlin Tasker y Evan Twomey Fotógrafo indicado debajo de cada foto. Producido por: Tyana Wachter, R. Foster, J. Philipp. Con el apoyo de Connie Keller, el Keller Action Science Center y el Andrew W. Mellon Foundation. © Kaitlin Tasker [[email protected]] y Evan Twomey [[email protected]] y William Duellman [[email protected]] (M): Macho, (H): Hembra, (Juv.): Juvenil © Science and Education, The Field Museum, Chicago [[email protected]] [fieldguides.fieldmuseum.org] Rapid Color Guide # 596 versión 1 10/2015 1 Allobates femoralis 2 Allobates femoralis 3 Allobates ornatus 4 Allobates trilineatus AROMOBATIDAE AROMOBATIDAE AROMOBATIDAE AROMOBATIDAE (foto: K. Tasker) (foto: W. Duellman) (foto: W. Duellman) (foto: W. Duellman) 5 Atelopus pulcher 6 Atelopus pulcher 7 Rhinella cf. festae 8 Rhinella cf. festae BUFONIDAE BUFONIDAE BUFONIDAE BUFONIDAE (foto: K. Tasker) (foto: K. Tasker) (foto: K. Tasker) (foto: K. Tasker) 9 Rhinella cf. festae 10 Rhinella margaritifera 11 Rhinella margaritifera 12 Rhinella marina BUFONIDAE BUFONIDAE BUFONIDAE BUFONIDAE (foto: K. Tasker) (foto: K. Tasker) (foto: K. Tasker) 13 Rhinella marina 14 Rhinella poeppigii 15 Cochranella erminea 16 Hyalinobatrachium cf. carlesvilai BUFONIDAE BUFONIDAE CENTROLENIDAE CENTROLENIDAE (foto: K. Tasker) (foto: W. Duellman) (foto: E. Twomey) (foto: E. Twomey) 17 Hyalinobatrachium pellucidum 18 Hyalinobatrachium pellucidum 19 Hyalinobatrachium pellucidum 20 Nymphargus chancas CENTROLENIDAE CENTROLENIDAE CENTROLENIDAE CENTROLENIDAE (foto: W. Duellman) (foto: W. Duellman) (foto: K. Tasker) (foto: E. Twomey) Parque Nacional Cordillera Azul, San Martín, Loreto, Ucayali y Huánuco, PERÚ 2 ANFIBIOS de Cordillera Azul Kaitlin Tasker y Evan Twomey Fotógrafo indicado debajo de cada foto. -

Volume and Geographical Distribution of Ecological Research in the Andes and the Amazon, 1995-2008
Mongabay.com Open Access Journal - Tropical Conservation Science Vol.4 (1):64-81, 2011 Research Article Volume and Geographical Distribution of Ecological Research in the Andes and the Amazon, 1995-2008 1,* 2 3 Nigel C. A. Pitman , Jocelyn Widmer , Clinton N. Jenkins , 4 5 5 Gabriela Stocks , Lisa Seales , Franklin Paniagua , Emilio 6 M. Bruna 1Amazon Conservation Association, Jirón Cusco 499, Puerto Maldonado, Madre de Dios, Peru. Current address: Center for Tropical Conservation, Nicholas School of the Environment, Box 90381, Duke University, Durham, NC 27708-0381 USA 2Department of Urban and Regional Planning, University of Florida, P. O. Box 115701, Gainesville, FL 32611-5701 USA 3Department of Biology, 1210 Biology-Psychology Building, University of Maryland, College Park, MD 20742 USA 4Department of Anthropology & Land Use and Environmental Change Institute, University of Florida, P.O. Box 117305, Gainesville, FL 32611-7305 USA 5The School of Natural Resources and Environment, University of Florida, P. O. Box 116455, Gainesville, FL 32611- 6455 USA 6Dept. of Wildlife Ecology & Conservation and Center for Latin American Studies, University of Florida, P.O. Box 110430, Gainesville, FL 32611-0430 USA *Corresponding author; email: [email protected] Received: 16 August 2010; Accepted: 10 January 2011; Published: 28 March 2011. Copyright: © Nigel C. A. Pitman, Jocelyn Widmer, Clinton N. Jenkins, Gabriela Stocks, Lisa Seales, Franklin Paniagua, Emilio M. Bruna. This is an open access paper. We use the Creative Commons Attribution 3.0 license http://creativecommons.org/licenses/by/3.0/ - The license permits any user to download, print out, extract, archive, and distribute the article, so long as appropriate credit is given to the authors and source of the work.