Does Batrachotoxin Autoresistance Co-Evolve with Toxicity in Phyllobates Poison-Dart Frogs?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Article Look but Don't Lick! Find out How These Brightly Colored Frogs
The Power of Poison Look But Don’t Lick! GRADES K-2 NOTES FOR EDUCATORS Common Core State Standards: W.K-2.2, W.K-2.8 Have students read the article “Look but Don’t Lick!” Have them write notes RI.K-2.1, RI.K-2.2, RI.K-2.4, RI.K-2.7, RI.K-2.10 in the large right-hand margin. For example, they could underline key passages, paraphrase important information, or write down questions that New York State Science Core Curriculum: they have. LE 3.1a Next Generation Science Standards: If it is not possible to create color handouts, use a computer projector to PE 1-LS1-2 display the reading so that students can see the colorful frog photos. You DCI LS1.A: Structure and Function may also have them color their black and white copies to match the actual All organisms have external parts. Different colors. animals use their body parts in different ways to see, hear, grasp objects, protect Ask: themselves, move from place to place, and • What does it mean for an animal to be poisonous? (A: An animal is seek, find, and take in food, water and air. poisonous if its body contains a substance that is harmful or fatal to other Plants also have different parts (roots, animals.) stems, leaves, flowers, fruits) that help • How does being poisonous help the frogs in this article survive? (A: them survive and grow. Predators will not eat an animal that is poisonous to them. The frogs signal that they are poisonous by their bright colors, which warn predators not to eat them. -

Catalogue of the Amphibians of Venezuela: Illustrated and Annotated Species List, Distribution, and Conservation 1,2César L
Mannophryne vulcano, Male carrying tadpoles. El Ávila (Parque Nacional Guairarepano), Distrito Federal. Photo: Jose Vieira. We want to dedicate this work to some outstanding individuals who encouraged us, directly or indirectly, and are no longer with us. They were colleagues and close friends, and their friendship will remain for years to come. César Molina Rodríguez (1960–2015) Erik Arrieta Márquez (1978–2008) Jose Ayarzagüena Sanz (1952–2011) Saúl Gutiérrez Eljuri (1960–2012) Juan Rivero (1923–2014) Luis Scott (1948–2011) Marco Natera Mumaw (1972–2010) Official journal website: Amphibian & Reptile Conservation amphibian-reptile-conservation.org 13(1) [Special Section]: 1–198 (e180). Catalogue of the amphibians of Venezuela: Illustrated and annotated species list, distribution, and conservation 1,2César L. Barrio-Amorós, 3,4Fernando J. M. Rojas-Runjaic, and 5J. Celsa Señaris 1Fundación AndígenA, Apartado Postal 210, Mérida, VENEZUELA 2Current address: Doc Frog Expeditions, Uvita de Osa, COSTA RICA 3Fundación La Salle de Ciencias Naturales, Museo de Historia Natural La Salle, Apartado Postal 1930, Caracas 1010-A, VENEZUELA 4Current address: Pontifícia Universidade Católica do Río Grande do Sul (PUCRS), Laboratório de Sistemática de Vertebrados, Av. Ipiranga 6681, Porto Alegre, RS 90619–900, BRAZIL 5Instituto Venezolano de Investigaciones Científicas, Altos de Pipe, apartado 20632, Caracas 1020, VENEZUELA Abstract.—Presented is an annotated checklist of the amphibians of Venezuela, current as of December 2018. The last comprehensive list (Barrio-Amorós 2009c) included a total of 333 species, while the current catalogue lists 387 species (370 anurans, 10 caecilians, and seven salamanders), including 28 species not yet described or properly identified. Fifty species and four genera are added to the previous list, 25 species are deleted, and 47 experienced nomenclatural changes. -

The Tadpole of Adelphobates Galactonotus (Steindachner, 1864) (Amphibia, Anura, Dendrobatidae)
Zootaxa 4422 (2): 287–290 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Correspondence ZOOTAXA Copyright © 2018 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4422.2.8 http://zoobank.org/urn:lsid:zoobank.org:pub:C16E5D1D-D75C-4411-94AE-41C038CA7BA9 The tadpole of Adelphobates galactonotus (Steindachner, 1864) (Amphibia, Anura, Dendrobatidae) DANUSY LOPES SANTOS1,2, 3, SILIONAMÃ PEREIRA DANTAS4 & FAUSTO NOMURA1 1Laboratório de Herpetologia e Comportamento Animal, Departamento de Ecologia, Universidade Federal de Goiás. Campus Samambaia, CEP 74001-970, Goiânia, GO, Brazil 2Laboratório de Ecologia Teórica, Departamento de Zoologia e Botânica, Universidade Estadual Paulista, Campus São José do Rio Preto. CEP 15054-000, São José do Rio Preto, São Paulo, Brazil 4Universidade Federal de Tocantins, Campus Araguaína, CEP 77838-824, Araguaína, TO, Brazil 3Corresponding author. E-mail: [email protected] The systematics of the dart-poison frogs, family Dendrobatidae, experienced several taxonomic rearrangements over time (e.g., Grant et al. 2006, 2017; Brown et al. 2011). Currently, this family comprises 194 described species organized in three sub-families and 15 genera (Frost 2018). Among them, the genus Adelphobates Grant, Frost, Caldwell, Gagliardo, Haddad, Kok, Means, Noonan, Schargel, & Wheeler, 2006, comprises three species, all distributed in Central and lower Amazon drainage of Peru and Brazil, and possibly in northeast of Bolivia (Grant et al. 2006; Frost 2018). Adelphobates galactonotus (Steindachner 1864) is an endemic Brazilian frog, and can be found throughout Pará, Maranhão, Mato Grosso and Tocantins states (Hoogmoed & Avila-Pires 2012), related to Amazon forest formations and also in transitional areas between the Cerrado and the Amazon forest (Valdujo et al. -

AC26 Doc. 20 Annex 1 English Only / Únicamente En Inglés / Seulement En Anglais
AC26 Doc. 20 Annex 1 English only / únicamente en inglés / seulement en anglais Fauna: new species and other taxonomic changes relating to species listed in the EC wildlife trade regulations January, 2012 A report to the European Commission Directorate General E - Environment ENV.E.2. – Environmental Agreements and Trade by the United Nations Environment Programme World Conservation Monitoring Centre AC26 Doc. 20, Annex 1 UNEP World Conservation Monitoring Centre 219 Huntingdon Road Cambridge CB3 0DL United Kingdom Tel: +44 (0) 1223 277314 Fax: +44 (0) 1223 277136 Email: [email protected] Website: www.unep-wcmc.org CITATION ABOUT UNEP-WORLD CONSERVATION UNEP-WCMC. 2012. Fauna: new species and MONITORING CENTRE other taxonomic changes relating to species The UNEP World Conservation Monitoring listed in the EC wildlife trade regulations. A Centre (UNEP-WCMC), based in Cambridge, report to the European Commission. UNEP- UK, is the specialist biodiversity information WCMC, Cambridge. and assessment centre of the United Nations Environment Programme (UNEP), run PREPARED FOR cooperatively with WCMC, a UK charity. The Centre's mission is to evaluate and highlight The European Commission, Brussels, Belgium the many values of biodiversity and put authoritative biodiversity knowledge at the DISCLAIMER centre of decision-making. Through the analysis and synthesis of global biodiversity The contents of this report do not necessarily knowledge the Centre provides authoritative, reflect the views or policies of UNEP or strategic and timely information for contributory organisations. The designations conventions, countries and organisations to use employed and the presentations do not imply in the development and implementation of the expressions of any opinion whatsoever on their policies and decisions. -

Species Diversity and Conservation Status of Amphibians in Madre De Dios, Southern Peru
Herpetological Conservation and Biology 4(1):14-29 Submitted: 18 December 2007; Accepted: 4 August 2008 SPECIES DIVERSITY AND CONSERVATION STATUS OF AMPHIBIANS IN MADRE DE DIOS, SOUTHERN PERU 1,2 3 4,5 RUDOLF VON MAY , KAREN SIU-TING , JENNIFER M. JACOBS , MARGARITA MEDINA- 3 6 3,7 1 MÜLLER , GIUSEPPE GAGLIARDI , LILY O. RODRÍGUEZ , AND MAUREEN A. DONNELLY 1 Department of Biological Sciences, Florida International University, 11200 SW 8th Street, OE-167, Miami, Florida 33199, USA 2 Corresponding author, e-mail: [email protected] 3 Departamento de Herpetología, Museo de Historia Natural de la Universidad Nacional Mayor de San Marcos, Avenida Arenales 1256, Lima 11, Perú 4 Department of Biology, San Francisco State University, 1600 Holloway Avenue, San Francisco, California 94132, USA 5 Department of Entomology, California Academy of Sciences, 55 Music Concourse Drive, San Francisco, California 94118, USA 6 Departamento de Herpetología, Museo de Zoología de la Universidad Nacional de la Amazonía Peruana, Pebas 5ta cuadra, Iquitos, Perú 7 Programa de Desarrollo Rural Sostenible, Cooperación Técnica Alemana – GTZ, Calle Diecisiete 355, Lima 27, Perú ABSTRACT.—This study focuses on amphibian species diversity in the lowland Amazonian rainforest of southern Peru, and on the importance of protected and non-protected areas for maintaining amphibian assemblages in this region. We compared species lists from nine sites in the Madre de Dios region, five of which are in nationally recognized protected areas and four are outside the country’s protected area system. Los Amigos, occurring outside the protected area system, is the most species-rich locality included in our comparison. -

Thermal Adaptation of Amphibians in Tropical Mountains
Thermal adaptation of amphibians in tropical mountains. Consequences of global warming Adaptaciones térmicas de anfibios en montañas tropicales: consecuencias del calentamiento global Adaptacions tèrmiques d'amfibis en muntanyes tropicals: conseqüències de l'escalfament global Pol Pintanel Costa ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel·lectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. -

Advertisement Call of Pratt's Rocket Frog, Colostethus Pratti, from The
SALAMANDRA 56(4): 395–400 SALAMANDRA 30 October 2020 ISSN 0036–3375 Correspondence German Journal of Herpetology Correspondence Advertisement call of Pratt’s rocket frog, Colostethus pratti, from the western Andes of Colombia (Anura: Dendrobatidae) Juan David Jiménez-Bolaño1, Andrés Camilo Montes-Correa1, Franziska Leonhardt2 & Juan Manuel Renjifo1 1) Grupo de Investigación en Manejo & Conservación de Fauna, Flora & Ecosistemas Estratégicos Neotropicales (MIKU), Universidad del Magdalena, Santa Marta, Colombia 2) Museum of Zoology, Senckenberg Natural History Collections Dresden, A. B. Meyer Building, 01109 Dresden, Germany Corresponding author: Andrés Camilo Montes-Correa, e-mail: [email protected] Manuscript received: 22 July 2019 Accepted: 14 April 2020 by Stefan Lötters Northwestern South America is one of the biologically most phibian checklists (Cochran & Goin 1970, Ruiz-Car- diverse yet least known regions of the world. The Tropi- ranza et al. 1994, Acosta-Galvis 2000, Lynch & Suá- cal Andes and Chocó-Tumbes-Magdalena ecoregions, two rez-Mayorga 2004, Ballesteros-Correa et al. 2019). hotspots with high degrees of endemism and conservation The most recent information onC. pratti in Colombia con- priority (Myers et al. 2000, Rodríguez-Mahecha et al. cerns its altitudinal distribution, relative abundance, and 2004), are part of this region. Especially the Chocoan low- natural history in the northernmost part of the western lands and the western Andes of Colombia are areas with Andes, at the Serranía de San Jerónimo, Departamento de exceptional high amphibian diversity and many endemic Córdoba (Romero-Martínez et al. 2008, Ballesteros- taxa (e.g., Lynch et al. 1997, Lynch & Suárez-Mayorga Correa et al. 2019). To the best of our knowledge, there is 2004, Armesto & Señaris 2017). -

N.Orntates PUBLISHED by the AMERICAN MUSEUM of NATURAL HISTORY CENTRAL PARK WEST at 79TH STREET, NEW YORK, N.Y
AMERICAN MUSEUM N.orntates PUBLISHED BY THE AMERICAN MUSEUM OF NATURAL HISTORY CENTRAL PARK WEST AT 79TH STREET, NEW YORK, N.Y. 10024 Number 3068, 15 pp., 12 figures, 1 table June 11, 1993 A New Poison Frog from Manu INational Park, Southeastern Peru (Dendrobatidae, Epipedobates) LILY RODRIGUEZ' AND CHARLES W. MYERS2 ABSTRACT Epipedobates macero is a new species of den- ilar to a few other species occurring along the An- drobatid poison frog from lowland rain forest of dean front in eastern Peru, namely E. petersi and the Manu National Park, in the upper Madre de E. cainarachi, which differ in details ofcoloration, Dios drainage ofsoutheastern Peru. It is most sim- morphology, and vocalization. RESUMEN Epipedobates macero, especie nueva, es un den- del llano amaz6nico al pie de los Andes orientales drobatido venenoso de la selva pluvial baja del peruanos, a saber, E. petersi y E. cainarachi, las Parque Nacional del Manu, en el drenaje del Rio cuales difieren en detalles de coloracion, morfo- Alto Madre de Dios, al sudeste del Peru. Es similar logla, y vocalizacion. a otras dos especies que ocurren en los bosques ' Field Associate, Department of Herpetology and Ichthyology, American Museum of Natural History. Investi- gadora Asociada: Asociaci6n Peruana para la Conservaci6n de la Naturaleza (APECO), Parque Jos6 de Acosta 187, Lima 17, Perfi; and Museo de Historia Natural de la Universidad Mayor de San Marcos, apartado 140434, Lima 14, Peru. 2 Curator, Department of Herpetology and Ichthyology, American Museum of Natural History. Copyright © American Museum of Natural History 1993 ISSN 0003-0082 / Price $3.90 2 AMERICAN MUSEUM NOVITATES NO. -

Chromosome Analysis of Five Brazilian
c Indian Academy of Sciences RESEARCH ARTICLE Chromosome analysis of five Brazilian species of poison frogs (Anura: Dendrobatidae) PAULA CAMARGO RODRIGUES1, ODAIR AGUIAR2, FLÁVIA SERPIERI1, ALBERTINA PIMENTEL LIMA3, MASAO UETANEBARO4 and SHIRLEI MARIA RECCO-PIMENTEL1∗ 1Departamento de Anatomia, Biologia Celular e Fisiologia, Instituto de Biologia, Universidade Estadual de Campinas, 13083-863 Campinas, São Paulo, Brazil 2Departamento de Biociências, Universidade Federal de São Paulo, Campus Baixada Santista, 11060-001 Santos, São Paulo, Brazil 3Coordenadoria de Pesquisas em Ecologia, Instituto Nacional de Pesquisas do Amazonas, 69011-970 Manaus, Amazonas, Brazil 4Departamento de Biologia, Universidade Federal de Mato Grosso do Sul, 70070-900 Campo Grande, Mato Grosso do Sul, Brazil Abstract Dendrobatid frogs have undergone an extensive systematic reorganization based on recent molecular findings. The present work describes karyotypes of the Brazilian species Adelphobates castaneoticus, A. quinquevittatus, Ameerega picta, A. galactonotus and Dendrobates tinctorius which were compared to each other and with previously described related species. All karyotypes consisted of 2n = 18 chromosomes, except for A. picta which had 2n = 24. The karyotypes of the Adelphobates and D. tinctorius species were highly similar to each other and to the other 2n = 18 previously studied species, revealing conserved karyotypic characteristics in both genera. In recent phylogenetic studies, all Adelphobates species were grouped in a clade separated from the Dendrobates species. Thus, we hypothesized that their common karyotypic traits may have a distinct origin by chromosome rearrangements and mutations. In A. picta, with 2n = 24, chromosome features of pairs from 1 to 8 are shared with other previously karyotyped species within this genus. Hence, the A. -
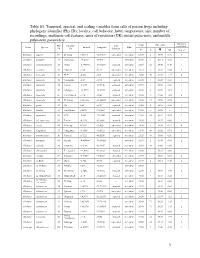
1 Table S1. Temporal, Spectral, and Scaling Variables from Calls Of
Table S1. Temporal, spectral, and scaling variables from calls of poison frogs including phylogeny identifier (Phy ID), locality, call behavior, habit, temperature, size, number of recordings, multinote call features, units of repetition (UR), initial pulse-note, and middle pulse-note parameters. Analyzed Phy Locality Call Temp SVL (mm) Genus Species Latitude Longitude Habit recordings ID ID Behavior °C N ! SD N of ♂ Allobates algorei 60 El Tama 7.65375 -72.19137 concealed terrestrial 23.50 8 18.90 0.70 3 Allobates brunneus 37 Guimaraes -15.2667 -55.5311 -- terrestrial 26.50 1 16.13 0.00 1 Allobates caeruleodactylus 48 Borba -4.398593 -59.60251 exposed terrestrial 25.60 12 15.50 0.40 1 Allobates crombiei 52 Altamira -3.65 -52.38 concealed terrestrial 24.10 2 18.10 0.04 2 Allobates femoralis 43 ECY -0.633 -76.5 concealed terrestrial 25.60 20 23.58 1.27 6 Allobates femoralis 46 Porongaba -8.67 -72.78 exposed terrestrial 25.00 1 25.38 0.00 1 Allobates femoralis 44 Leticia -4.2153 -69.9406 exposed terrestrial 25.50 1 20.90 0.00 1 Allobates femoralis 40 Albergue -12.8773 -71.3865 exposed terrestrial 26.00 6 21.98 2.18 1 Allobates femoralis 41 CAmazonico -12.6 -70.08 exposed terrestrial 26.00 12 22.43 1.06 4 Allobates femoralis 45 El Palmar 8.333333 -61.66667 concealed terrestrial 24.00 27 25.50 0.76 1 Allobates granti 49 FG 3.62 -53.17 exposed terrestrial 24.60 8 16.15 0.55 1 Allobates humilis 59 San Ramon 8.8678 -70.4861 concealed terrestrial 19.50 -- 21.80 -- 1 Allobates insperatus 54 ECY -0.633 -76.4005 exposed terrestrial 24.60 18 16.64 0.93 7 Allobates aff. -

Visual Signaling in Anuran Amphibians
.. Hödl, W. and Amezquita, A. (2001). Visual signaling in anuran amphibians. In: Anuran communication, (M.J. Ryan, ed.). .. Smithsonian lust. Press, Washington. Pp. 121-141. 10 WALTER HÖDL AND ADOLFO AMEZQUITA Visual Signaling in Anuran Amphibians lntroduction cation. social behavior, or natural history. visual signaling was either not considered or was treated as a minor subject Acoustic communication plays a fundamental role in an- (Wells 1977a, 1977b; Arak 1983; Duellman and Trueb 1986; uran reproduction and thus is involved in evolutionary Rand 1988; Halliday and Tejedo 1995; Stebbins and Cohen processes such as mate recognition. reproductive isolation. 1995; Sullivan et al. 1995). The most detailed review ofthe speciation. and character displacement (Wells 1977a. 1977b. subject is now more than 20 years old (Wells 1977b). Never- 1988;Rand 1988;Gerhardt and Schwartz 1995;Halliday and theless some authors have discussed the possible evolution- Tejedo 1995;Sullivan et al. 1995).Visual cues. however. have ary link between visual signaling and the reproductive ecol- been thought to function only during dose-range inter- ogy of species, such as reproduction associated with streams actions (Wells 1977c; Duellman and Trueb 1986). Visual sig- (Heyer et aI. 1990; Lindquist and Hetherington 1996. 1998; naling is predicted to be predominantly employed by diur- Hödl et al. 1997;Haddad and Giaretta 1999) or reproduction nal species at sites with an unobstructed view (Endler 1992). within feeding territories (Wells 1977c). Diurnality. however. is not common for the majority offrog Our aim in this review is (1) to propose a dassmcation of species. Thus vocalizations. which are highly efficient for reported behavioral patterns of visual signaling in frags; (2) communicating at night or in dense vegetation, are by far to describe the diversity of visual signals among living an- the best studied anuran signals (Duellman and Trueb 1986; uran taxa; and (3) to apply a comparative approach to explor- Fritzsch et aI. -

Anura: Dendrobatidae: Anomaloglossus) from the Orinoquian Rainforest, Southern Venezuela
TERMS OF USE This pdf is provided by Magnolia Press for private/research use. Commercial sale or deposition in a public library or website is prohibited. Zootaxa 2413: 37–50 (2010) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2010 · Magnolia Press ISSN 1175-5334 (online edition) A new dendrobatid frog (Anura: Dendrobatidae: Anomaloglossus) from the Orinoquian rainforest, southern Venezuela CÉSAR L. BARRIO-AMORÓS1,4, JUAN CARLOS SANTOS2 & OLGA JOVANOVIC3 1,4 Fundación AndígenA, Apartado postal 210, 5101-A Mérida, Venezuela 2University of Texas at Austin, Integrative Biology, 1 University Station C0930 Austin TX 78705, USA 3Division of Evolutionary Biology, Technical University of Braunschweig, Spielmannstr 8, 38106 Braunschweig, Germany 4Corresponding author. E-mail: [email protected]; [email protected] Abstract A new species of Anomaloglossus is described from the Venezuelan Guayana; it is the 21st described species of Anomaloglossus from the Guiana Shield, and the 15th from Venezuela. This species inhabits rainforest on granitic substrate on the northwestern edge of the Guiana Shield (Estado Amazonas, Venezuela). The new species is distinguished from congeners by sexual dimorphism, its unique male color pattern (including two bright orange parotoid marks and two orange paracloacal spots on dark brown background), call characteristics and other morphological features. Though to the new species is known only from the northwestern edge of the Guiana Shield, its distribution may be more extensive, as there are no significant biogeographic barriers isolating the type locality from the granitic lowlands of Venezuela. Key words: Amphibia, Dendrobatidae, Anomaloglossus, Venezuela, Guiana Shield Resumen Se describe una nueva especie de Anomaloglossus de la Guayana venezolana, que es la vigesimoprimera descrita del Escudo Guayanés, y la decimoquinta para Venezuela.