Sumakuru, a Deeply-Diverging New Genus of Lyssomanine Jumping
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Molecular Phylogeny, Divergence Times and Biogeography of Spiders of the Subfamily Euophryinae (Araneae: Salticidae) ⇑ Jun-Xia Zhang A, , Wayne P
Molecular Phylogenetics and Evolution 68 (2013) 81–92 Contents lists available at SciVerse ScienceDirect Molec ular Phylo genetics and Evolution journal homepage: www.elsevier.com/locate/ympev Molecular phylogeny, divergence times and biogeography of spiders of the subfamily Euophryinae (Araneae: Salticidae) ⇑ Jun-Xia Zhang a, , Wayne P. Maddison a,b a Department of Zoology, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 b Department of Botany and Beaty Biodiversity Museum, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 article info abstract Article history: We investigate phylogenetic relationships of the jumping spider subfamily Euophryinae, diverse in spe- Received 10 August 2012 cies and genera in both the Old World and New World. DNA sequence data of four gene regions (nuclear: Revised 17 February 2013 28S, Actin 5C; mitochondrial: 16S-ND1, COI) were collected from 263 jumping spider species. The molec- Accepted 13 March 2013 ular phylogeny obtained by Bayesian, likelihood and parsimony methods strongly supports the mono- Available online 28 March 2013 phyly of a Euophryinae re-delimited to include 85 genera. Diolenius and its relatives are shown to be euophryines. Euophryines from different continental regions generally form separate clades on the phy- Keywords: logeny, with few cases of mixture. Known fossils of jumping spiders were used to calibrate a divergence Phylogeny time analysis, which suggests most divergences of euophryines were after the Eocene. Given the diver- Temporal divergence Biogeography gence times, several intercontinental dispersal event sare required to explain the distribution of euophry- Intercontinental dispersal ines. Early transitions of continental distribution between the Old and New World may have been Euophryinae facilitated by the Antarctic land bridge, which euophryines may have been uniquely able to exploit Diolenius because of their apparent cold tolerance. -
Open PDF Printout of This File
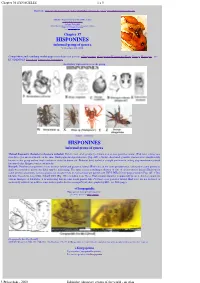
Chapter 36 SYNAGELES 1 z 5 Main links: Title page Introduction & guides INDEXES_chapters_&_genera Open PDF printout of this file Salticidae (Araneae) genera of the world - an atlas (unfinished manuscript) by Jerzy Prószy ński Professor Emeritus, Museum and Institute of Zoology, Polish Academy of Sciences, ul. Wilcza 63, 00-679 Warsaw, POLAND e-mail: [email protected] Chapter 37 HISPONINES informal group of genera Version June 3rd, 2020. symbol of the supragroup HISPONINES Composition and searching on this page (including fossil genera): +Gorgopsidis : +Gorgopsina Hermotimus Hispo Jerzego Massagris - see EUODENINES Tomobella Tomocyrba Tomomingi Exemplary representatives of the group Source - see below HISPONINES informal group of genera Mutual diagnostic characters of genera included . Differs from other groups by minute size of eyes posterior lateral (IInd row), sitting very closely to eyes anterior lateral, on the same black pigmented protuberance (Fig. 49J1). On the other hand, genitalic characters are insufficiently known in this group and no final conclusion could be drawn yet. Embolus bent, coiled or straight and minute, sitting atop membranous distal haematodocha. Epigyne varies extensively. Remark. Peculiar arrangement of eyes anterior lateral and posterior lateral (IInd row) on the same protuberance, followed in some genera by shallow constriction across eye field, may be misleading. The same concerns striking reduction of size of eyes posterior lateral (IInd row) in some genera classified to various groups, for instance eyes in Lystrocteissa are paired with HYLLINES[?] like palp structure (Figs 14L, 17G), likewise Tomobella fotsy Szüts, Scharff 2009 (Fig. 49I) resembles some Neon . That somatic character is apparently prone to develop parallel in various lineages of Salticidae, it is interesting that in some fossil genera (like Prolinus ) eyes posterior lateral (IInd row) are not reduced, or moderately reduced (as in Hisponinae amber spider shown on magnificent photographs by Hill - see Title page). -

Diversity of Simonid Spiders (Araneae: Salticidae: Salticinae) in India
IJBI 2 (2), (DECEMBER 2020) 247-276 International Journal of Biological Innovations Available online: http://ijbi.org.in | http://www.gesa.org.in/journals.php DOI: https://doi.org/10.46505/IJBI.2020.2223 Review Article E-ISSN: 2582-1032 DIVERSITY OF SIMONID SPIDERS (ARANEAE: SALTICIDAE: SALTICINAE) IN INDIA Rajendra Singh1*, Garima Singh2, Bindra Bihari Singh3 1Department of Zoology, Deendayal Upadhyay University of Gorakhpur (U.P.), India 2Department of Zoology, University of Rajasthan, Jaipur (Rajasthan), India 3Department of Agricultural Entomology, Janta Mahavidyalaya, Ajitmal, Auraiya (U.P.), India *Corresponding author: [email protected] Received: 01.09.2020 Accepted: 30.09.2020 Published: 09.10.2020 Abstract: Distribution of spiders belonging to 4 tribes of clade Simonida (Salticinae: Salticidae: Araneae) reported in India is dealt. The tribe Aelurillini (7 genera, 27 species) is represented in 16 states and in 2 union territories, Euophryini (10 genera, 16 species) in 14 states and in 4 union territories, Leptorchestini (2 genera, 3 species) only in 2 union territories, Plexippini (22 genera, 73 species) in all states except Mizoram and in 3 union territories, and Salticini (3 genera, 11 species) in 15 states and in 4 union terrioties. West Bengal harbours maximum number of species, followed by Tamil Nadu and Maharashtra. Out of 129 species of the spiders listed, 70 species (54.3%) are endemic to India. Keywords: Aelurillini, Euophryini, India, Leptorchestini, Plexippini, Salticidae, Simonida. INTRODUCTION Hisponinae, Lyssomaninae, Onomastinae, Spiders are chelicerate arthropods belonging to Salticinae and Spartaeinae. Out of all the order Araneae of class Arachnida. Till to date subfamilies, Salticinae comprises 93.7% of the 48,804 described species under 4,180 genera and species (5818 species, 576 genera, including few 128 families (WSC, 2020). -

Zootaxa 1255: 37–55 (2006) ISSN 1175-5326 (Print Edition) ZOOTAXA 1255 Copyright © 2006 Magnolia Press ISSN 1175-5334 (Online Edition)
Zootaxa 1255: 37–55 (2006) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA 1255 Copyright © 2006 Magnolia Press ISSN 1175-5334 (online edition) Lapsiines and hisponines as phylogenetically basal salticid spiders (Araneae: Salticidae) WAYNE P. MADDISON1 & KAREN M. NEEDHAM2 1Departments of Zoology and Botany and Cent re for Biod iversity Resea rch, University of Bri tish Columbia, 6270 University Bouleva rd, Vancouve r, British Columbia , V6T 1Z4, Canada. 2Spencer Entomological Museum , Department of Zoology, Univers ity of British Columbia, 6270 University Boulevard, Vancouve r, British Co lumbia , V6T 1Z4, Can Abstract Increased phylogenetic resolution of the basal lineages of salticid spiders will help us understand their early evolution and provide better outgroups for phylogenetic studies within the major clades. We gathered sequences of nuclear and mitochondrial gene regions (28S, 18S, Histone 3, 16S-ND1, CO1) and used them to reconstruct salticid phylogeny by parsimony, likelihood and Bayesian methods. Our results confirm that lapsiines and hisponines are among the basal salticids, i.e. outside the major clade Salticoida. The lapsiines are resolved as sister group to the spartaeines. The precise placement of hisponines is unclear, but they may represent a deep-branching lineage independent from the spartaeines. Key words: Araneae, Salticidae, Thrandin a, Galianora, Hispo, Massagris, Tomocyrb a, Goleba, lapsiines, Hisponinae, Spartaeinae, Lyssomaninae, jumping spider, basal groups, phylogeny Introduction Morphological and molecular data have begun to resolve the basal phylogenetic structure of salticid spiders (Wanless, 1980, 1982, 1984, Rodrigo & Jackson, 1992, Maddison, 1988, 1996, Wijesinghe, 1992, 1997, Maddison & Hedin, 2003). One of the best corroborated clades is the Salticoida (Maddison & Hedin, 2003), within which falls the vast majority of salticids, about 95% of the approximately 5000 described species (Platnick, 2005). -

Jerzego, a New Hisponine Jumping Spider from Borneo (Araneae: Salticidae)
Zootaxa 3852 (5): 569–578 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3852.5.5 http://zoobank.org/urn:lsid:zoobank.org:pub:85C58188-05CE-4EED-A371-8AFE0E8148C8 Jerzego, a new hisponine jumping spider from Borneo (Araneae: Salticidae) WAYNE P. MADDISON1,3 & EDYTA K. PIASCIK2 1Departments of Zoology and Botany and Beaty Biodiversity Museum, University of British Columbia, 6270 University Boulevard, Vancouver, British Columbia, V6T 1Z4, Canada. E-mail: [email protected] 2Department of Zoology, University of British Columbia, 6270 University Boulevard, Vancouver, British Columbia, V6T 1Z4, Canada. E-mail: [email protected] 3Corresponding author Abstract A new genus and species of hisponine jumping spider from Sarawak, Jerzego corticicola Maddison sp. nov. are described, representing one of the few hisponine jumping spiders known from Asia, and the only whose male is known. Although similar to the primarily-Madagascan genus Hispo in having an elongate and flat body, sequences of 28s and 16sND1 genes indicate that Jerzego is most closely related to Massagris and Tomomingi, a result consistent with morphology. Females of Jerzego and other genera of Hisponinae were found to have an unusual double copulatory duct, which appears to be a synapomorphy of the subfamily. Two species are transferred from Hispo, Jerzego bipartitus (Simon) comb. nov. and Jer- zego alboguttatus (Simon) comb. nov. Diagnostic illustrations -

Visual Perception in Jumping Spiders (Araneae,Salticidae)
Visual Perception in Jumping Spiders (Araneae,Salticidae) A thesis submitted in partial fulfilment of the requirements for the Degree of Doctor of Philosophy in Biology at the University of Canterbury by Yinnon Dolev University of Canterbury 2016 Table of Contents Abstract.............................................................................................................................................................................. i Acknowledgments .......................................................................................................................................................... iii Preface ............................................................................................................................................................................. vi Chapter 1: Introduction ................................................................................................................................................... 1 Chapter 2: Innate pattern recognition and categorisation in a jumping Spider ........................................................... 9 Abstract ....................................................................................................................................................................... 10 Introduction ................................................................................................................................................................ 11 Methods ..................................................................................................................................................................... -

Predatory Behavior of Jumping Spiders
Annual Reviews www.annualreviews.org/aronline Annu Rev. Entomol. 19%. 41:287-308 Copyrighl8 1996 by Annual Reviews Inc. All rights reserved PREDATORY BEHAVIOR OF JUMPING SPIDERS R. R. Jackson and S. D. Pollard Department of Zoology, University of Canterbury, Christchurch, New Zealand KEY WORDS: salticids, salticid eyes, Portia, predatory versatility, aggressive mimicry ABSTRACT Salticids, the largest family of spiders, have unique eyes, acute vision, and elaborate vision-mediated predatory behavior, which is more pronounced than in any other spider group. Diverse predatory strategies have evolved, including araneophagy,aggressive mimicry, myrmicophagy ,and prey-specific preycatch- ing behavior. Salticids are also distinctive for development of behavioral flexi- bility, including conditional predatory strategies, the use of trial-and-error to solve predatory problems, and the undertaking of detours to reach prey. Predatory behavior of araneophagic salticids has undergone local adaptation to local prey, and there is evidence of predator-prey coevolution. Trade-offs between mating and predatory strategies appear to be important in ant-mimicking and araneo- phagic species. INTRODUCTION With over 4000 described species (1 l), jumping spiders (Salticidae) compose by Fordham University on 04/13/13. For personal use only. the largest family of spiders. They are characterized as cursorial, diurnal predators with excellent eyesight. Although spider eyes usually lack the struc- tural complexity required for acute vision, salticids have unique, complex eyes with resolution abilities without known parallels in animals of comparable size Annu. Rev. Entomol. 1996.41:287-308. Downloaded from www.annualreviews.org (98). Salticids are the end-product of an evolutionary process in which a small silk-producing animal with a simple nervous system acquires acute vision, resulting in a diverse array of complex predatory strategies. -

Two New Genera of Jumping Spiders from Hainan Island, China (Araneae, Salticidae)
Zootaxa 3712 (1): 001–084 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Monograph ZOOTAXA Copyright © 2013 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3712.1.1 http://zoobank.org/urn:lsid:zoobank.org:pub:91ACA25B-A016-40ED-B105-3D9D960CA92E ZOOTAXA 3712 Two New Genera of Jumping Spiders from Hainan Island, China (Araneae, Salticidae) YUANYE ZHOU 1, 2 & SHUQIANG LI 1, 3 1Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China 2Liaoning Key Laboratory of Evolution and Biodiversity, Shenyang Normal University, Shenyang 110034, China 3 Corresponding author. E-mail: [email protected] Magnolia Press Auckland, New Zealand Accepted by T. Szuts: 8 Aug. 2013; published: 19 Sept. 2013 YUANYE ZHOU & SHUQIANG LI Two New Genera of Jumping Spiders from Hainan Island, China (Araneae, Salticidae) (Zootaxa 3712) 84 pp.; 30 cm. 19 Sept. 2013 ISBN 978-1-77557-262-6 (paperback) ISBN 978-1-77557-263-3 (Online edition) FIRST PUBLISHED IN 2013 BY Magnolia Press P.O. Box 41-383 Auckland 1346 New Zealand e-mail: [email protected] http://www.mapress.com/zootaxa/ © 2013 Magnolia Press All rights reserved. No part of this publication may be reproduced, stored, transmitted or disseminated, in any form, or by any means, without prior written permission from the publisher, to whom all requests to reproduce copyright material should be directed in writing. This authorization does not extend to any other kind of copying, by any means, in any form, and for any purpose other than private research use. ISSN 1175-5326 (Print edition) ISSN 1175-5334 (Online edition) 2 · Zootaxa 3712 (1) © 2013 Magnolia Press ZHOU & LI Table of contents Abstract . -

(Arachnida: Araneae) of the Floodplain Forests of the Main Amazon River Channel
ARTÍCULO: A contribution to the knowledge of the spider fauna (Arachnida: Araneae) of the floodplain forests of the main Amazon River channel Felipe N. A. A. Rego, Eduardo M. Venticinque, Antonio D. Brescovit, Cristina A. Rheims & Ana L. K. M. Albernaz Abstract: ARTÍCULO: We collected spiders during an expedition along 3000 km of the floodplains of the Brazilian part of the main channel of the Amazon River and identified them A contribution to the knowledge of to family, genus and species / morphospecies level whenever possible. More the spider fauna (Arachnida: Ara- than half of the collected species represented new records. The percentage of neae) of the floodplain forests of the singletons (35.6%) and doubletons (17.4%), the lack of overlapping between main Amazon River channel the data obtained in this study and that of the literature, and the under sampling Felipe N. A. A. Rego emphasizes the need for more inventories in the Amazon River floodplain and Pós-Graduação em Ecologia, Univer- a more complete set of sampling methods, such as canopy fogging and pitfall sidade de Brasília, 70919-970, Brasí- trapping. Therefore, knowledge on the fauna of the Amazon floodplains will lia, DF, Brazil. [email protected] remain an enormous challenge, regarding the still superficial collecting efforts, Eduardo M. Venticinque the lack of long-term samplings, taxonomic knowledge and capacity. Wildlife Conser. Soc., Rua dos Jato- Key words: Arachnida, Araneae, spiders, inventory, Amazon River, várzea, Amazo- bás, 274, Coroado 3, 69085-000 and nia. INPA, 69011-970, C.P. 478, Manaus, AM, Brazil. [email protected] A. D. -

A Protocol for Online Documentation of Spider Biodiversity Inventories Applied to a Mexican Tropical Wet Forest (Araneae, Araneomorphae)
Zootaxa 4722 (3): 241–269 ISSN 1175-5326 (print edition) https://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2020 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4722.3.2 http://zoobank.org/urn:lsid:zoobank.org:pub:6AC6E70B-6E6A-4D46-9C8A-2260B929E471 A protocol for online documentation of spider biodiversity inventories applied to a Mexican tropical wet forest (Araneae, Araneomorphae) FERNANDO ÁLVAREZ-PADILLA1, 2, M. ANTONIO GALÁN-SÁNCHEZ1 & F. JAVIER SALGUEIRO- SEPÚLVEDA1 1Laboratorio de Aracnología, Facultad de Ciencias, Departamento de Biología Comparada, Universidad Nacional Autónoma de México, Circuito Exterior s/n, Colonia Copilco el Bajo. C. P. 04510. Del. Coyoacán, Ciudad de México, México. E-mail: [email protected] 2Corresponding author Abstract Spider community inventories have relatively well-established standardized collecting protocols. Such protocols set rules for the orderly acquisition of samples to estimate community parameters and to establish comparisons between areas. These methods have been tested worldwide, providing useful data for inventory planning and optimal sampling allocation efforts. The taxonomic counterpart of biodiversity inventories has received considerably less attention. Species lists and their relative abundances are the only link between the community parameters resulting from a biotic inventory and the biology of the species that live there. However, this connection is lost or speculative at best for species only partially identified (e. g., to genus but not to species). This link is particularly important for diverse tropical regions were many taxa are undescribed or little known such as spiders. One approach to this problem has been the development of biodiversity inventory websites that document the morphology of the species with digital images organized as standard views. -

The Jumping Spiders (Araneae: Salticidae) of the Virginia Peninsula1
Vol. 98, No. 5, November & December 1987 235 THE JUMPING SPIDERS (ARANEAE: SALTICIDAE) OF THE VIRGINIA PENINSULA 1 2 C.L. Stietenroth, N.V. Horner ABSTRACT: Thirty species representing 1 8 genera of Salticidae are recorded from the Virginia Peninsula. Habitat and natural history information for each species is presented. Some salticids on the peninsula occupy diverse habitats while other species appear to confine themselves to more restricted environments. The most abundant salticid was Hentzia palmarum. Metaphi- dippus galathea and Platycryptus undatus were most widely distributed species. Salticids reported in Virginia for the first time are Phidippus princeps, P. otiosus, Thiodina sylvana, Sitticus fasciger and Zygoballus sexpunctatus. A few studies concerning the spider fauna of Virginia have been published. The earliest record of occurrence was by John Banister between 1678 and 1692 (Ewan and Ewan, 1970). More recently, McCaffrey and Hornsburgh published three studies concerning spiders in apple orchards in central Virginia. Their assessment of spider populations in an unsprayed orchard was published in 1 1 977 followed ( 978) by laboratory feeding studies performed to evaluate potential effects of predaceous spiders on insect residents of apple orchards. Later (1980), a comparison was made between the spider populations in abandoned and commercial orchards; 68 species were identified. Dowd and Kok (1981), and McPherson el al. (1982) considered spider and other arthropod predation on the curculionid beetle, Rhynocyllus sp., in a in 1 soybean cropping system Virginia. Holsinger ( 982) reported on the spider cave-fauna in Burnsville Cove. The efficiency of limb-beating for capturing various spider families in apple orchards is discussed by McCaffrey and Parrella(1984). -

Jahresbericht 2018 Der Generaldirektion Der Staatlichen Naturwissenschaftlichen Sammlungen Bayerns Herausgegeben Von: Prof
Jahresbericht 2018 der Generaldirektion der Staatlichen Naturwissenschaftlichen Sammlungen Bayerns Herausgegeben von: Prof. Dr. Gerhard Haszprunar, Generaldirektor Generaldirektion der Staatlichen Naturwissenschaftlichen Sammlungen Bayerns (SNSB) Menzinger Straße 71, 80638 München erschienen: München im August 2019 Zusammenstellung und Endredaktion: Dr. Eva Maria Natzer (Generaldirektion) Helene Tobollik (Generaldirektion) Unterstützung durch: Katja Henßel (Generaldirektion) Druck: GC Digitaldruck Guido Coenen, München Inhaltsverzeichnis Bericht des Generaldirektors .......................................................................................................................4 Wissenschaftliche Publikationen ....................................................................................................................6 Projektmittelübersicht 2018 ...........................................................................................................................47 Organigramm der SNSB: ..............................................................................................................................59 Generaldirektion ..........................................................................................................................................60 Personalvertretung der SNSB .......................................................................................................................63 Museen: Geologisches Museum München ..................................................................................................................64