SUPPORTING INFORMATION APPENDIX Mitochondrial Genomes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Aberrant Modulation of Ribosomal Protein S6 Phosphorylation Confers Acquired Resistance to MAPK Pathway Inhibitors in BRAF-Mutant Melanoma
www.nature.com/aps ARTICLE Aberrant modulation of ribosomal protein S6 phosphorylation confers acquired resistance to MAPK pathway inhibitors in BRAF-mutant melanoma Ming-zhao Gao1,2, Hong-bin Wang1,2, Xiang-ling Chen1,2, Wen-ting Cao1,LiFu1, Yun Li1, Hai-tian Quan1,2, Cheng-ying Xie1,2 and Li-guang Lou1,2 BRAF and MEK inhibitors have shown remarkable clinical efficacy in BRAF-mutant melanoma; however, most patients develop resistance, which limits the clinical benefit of these agents. In this study, we found that the human melanoma cell clones, A375-DR and A375-TR, with acquired resistance to BRAF inhibitor dabrafenib and MEK inhibitor trametinib, were cross resistant to other MAPK pathway inhibitors. In these resistant cells, phosphorylation of ribosomal protein S6 (rpS6) but not phosphorylation of ERK or p90 ribosomal S6 kinase (RSK) were unable to be inhibited by MAPK pathway inhibitors. Notably, knockdown of rpS6 in these cells effectively downregulated G1 phase-related proteins, including RB, cyclin D1, and CDK6, induced cell cycle arrest, and inhibited proliferation, suggesting that aberrant modulation of rpS6 phosphorylation contributed to the acquired resistance. Interestingly, RSK inhibitor had little effect on rpS6 phosphorylation and cell proliferation in resistant cells, whereas P70S6K inhibitor showed stronger inhibitory effects on rpS6 phosphorylation and cell proliferation in resistant cells than in parental cells. Thus regulation of rpS6 phosphorylation, which is predominantly mediated by BRAF/MEK/ERK/RSK signaling in parental cells, was switched to mTOR/ P70S6K signaling in resistant cells. Furthermore, mTOR inhibitors alone overcame acquired resistance and rescued the sensitivity of the resistant cells when combined with BRAF/MEK inhibitors. -

Allele-Specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish
Allele-specific Expression of Ribosomal Protein Genes in Interspecific Hybrid Catfish by Ailu Chen A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama August 1, 2015 Keywords: catfish, interspecific hybrids, allele-specific expression, ribosomal protein Copyright 2015 by Ailu Chen Approved by Zhanjiang Liu, Chair, Professor, School of Fisheries, Aquaculture and Aquatic Sciences Nannan Liu, Professor, Entomology and Plant Pathology Eric Peatman, Associate Professor, School of Fisheries, Aquaculture and Aquatic Sciences Aaron M. Rashotte, Associate Professor, Biological Sciences Abstract Interspecific hybridization results in a vast reservoir of allelic variations, which may potentially contribute to phenotypical enhancement in the hybrids. Whether the allelic variations are related to the downstream phenotypic differences of interspecific hybrid is still an open question. The recently developed genome-wide allele-specific approaches that harness high- throughput sequencing technology allow direct quantification of allelic variations and gene expression patterns. In this work, I investigated allele-specific expression (ASE) pattern using RNA-Seq datasets generated from interspecific catfish hybrids. The objective of the study is to determine the ASE genes and pathways in which they are involved. Specifically, my study investigated ASE-SNPs, ASE-genes, parent-of-origins of ASE allele and how ASE would possibly contribute to heterosis. My data showed that ASE was operating in the interspecific catfish system. Of the 66,251 and 177,841 SNPs identified from the datasets of the liver and gill, 5,420 (8.2%) and 13,390 (7.5%) SNPs were identified as significant ASE-SNPs, respectively. -

Selective Induction of Apoptosis in HIV-Infected Macrophages
Selective Induction of Apoptosis in HIV-infected Macrophages by Simon Xin Min Dong A thesis submitted in partial fulfillment of the requirements for the Doctorate in Philosophy degree in Microbiology and Immunology Department of Biochemistry, Microbiology, and Immunology Faculty of Medicine University of Ottawa © Simon Xin Min Dong, Ottawa, Canada, 2020 Abstract The eradication of Human Immunodeficiency Virus (HIV) from infected patients is one of the major medical problems of our time, primarily due to HIV reservoir formation. Macrophages play important roles in HIV reservoir formation: once infected, they shield HIV against host anti-viral immune responses and anti-retroviral therapies, help viral spread and establish infection in anatomically protected sites. Thus, it is imperative to selectively induce the apoptosis of HIV-infected macrophages for a complete cure of this disease. I hypothesize that HIV infection dysregulates the expression of some specific genes, which is essential to the survival of infected host cells, and these genes can be targeted to selectively induce the apoptosis of HIV-infected macrophages. My objective is to identify the genes that can be targeted to eradicate HIV reservoir in macrophages, and to briefly elucidate the mechanism of cell death induced by targeting one of the identified genes. A four-step strategy was proposed to reach the goal. First, 90k shRNA lentivirus pool technology and microarray analysis were employed in a genome-wide screen of genes and 28 promising genes were found. Second, siRNA silencing was applied to validate these genes with 2 different HIV-1 viruses; as a result, 4 genes, Cox7a2, Znf484, Cdk2, and Cstf2t, were identified to be novel gene targets. -

Multi-Targeted Mechanisms Underlying the Endothelial Protective Effects of the Diabetic-Safe Sweetener Erythritol
Multi-Targeted Mechanisms Underlying the Endothelial Protective Effects of the Diabetic-Safe Sweetener Erythritol Danie¨lle M. P. H. J. Boesten1*., Alvin Berger2.¤, Peter de Cock3, Hua Dong4, Bruce D. Hammock4, Gertjan J. M. den Hartog1, Aalt Bast1 1 Department of Toxicology, Maastricht University, Maastricht, The Netherlands, 2 Global Food Research, Cargill, Wayzata, Minnesota, United States of America, 3 Cargill RandD Center Europe, Vilvoorde, Belgium, 4 Department of Entomology and UCD Comprehensive Cancer Center, University of California Davis, Davis, California, United States of America Abstract Diabetes is characterized by hyperglycemia and development of vascular pathology. Endothelial cell dysfunction is a starting point for pathogenesis of vascular complications in diabetes. We previously showed the polyol erythritol to be a hydroxyl radical scavenger preventing endothelial cell dysfunction onset in diabetic rats. To unravel mechanisms, other than scavenging of radicals, by which erythritol mediates this protective effect, we evaluated effects of erythritol in endothelial cells exposed to normal (7 mM) and high glucose (30 mM) or diabetic stressors (e.g. SIN-1) using targeted and transcriptomic approaches. This study demonstrates that erythritol (i.e. under non-diabetic conditions) has minimal effects on endothelial cells. However, under hyperglycemic conditions erythritol protected endothelial cells against cell death induced by diabetic stressors (i.e. high glucose and peroxynitrite). Also a number of harmful effects caused by high glucose, e.g. increased nitric oxide release, are reversed. Additionally, total transcriptome analysis indicated that biological processes which are differentially regulated due to high glucose are corrected by erythritol. We conclude that erythritol protects endothelial cells during high glucose conditions via effects on multiple targets. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

Gene Expression Profiling of Corpus Luteum Reveals the Importance Of
bioRxiv preprint doi: https://doi.org/10.1101/673558; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Gene expression profiling of corpus luteum reveals the 2 importance of immune system during early pregnancy in 3 domestic sheep. 4 Kisun Pokharel1, Jaana Peippo2 Melak Weldenegodguad1, Mervi Honkatukia2, Meng-Hua Li3*, Juha 5 Kantanen1* 6 1 Natural Resources Institute Finland (Luke), Jokioinen, Finland 7 2 Nordgen – The Nordic Genetic Resources Center, Ås, Norway 8 3 College of Animal Science and Technology, China Agriculture University, Beijing, China 9 * Correspondence: MHL, [email protected]; JK, [email protected] 10 Abstract: The majority of pregnancy loss in ruminants occurs during the preimplantation stage, which is thus 11 the most critical period determining reproductive success. While ovulation rate is the major determinant of 12 litter size in sheep, interactions among the conceptus, corpus luteum and endometrium are essential for 13 pregnancy success. Here, we performed a comparative transcriptome study by sequencing total mRNA from 14 corpus luteum (CL) collected during the preimplantation stage of pregnancy in Finnsheep, Texel and F1 15 crosses, and mapping the RNA-Seq reads to the latest Rambouillet reference genome. A total of 21,287 genes 16 were expressed in our dataset. Highly expressed autosomal genes in the CL were associated with biological 17 processes such as progesterone formation (STAR, CYP11A1, and HSD3B1) and embryo implantation (eg. -

Screening and Identification of Hub Genes in Bladder Cancer by Bioinformatics Analysis and KIF11 Is a Potential Prognostic Biomarker
ONCOLOGY LETTERS 21: 205, 2021 Screening and identification of hub genes in bladder cancer by bioinformatics analysis and KIF11 is a potential prognostic biomarker XIAO‑CONG MO1,2*, ZI‑TONG ZHANG1,3*, MENG‑JIA SONG1,2, ZI‑QI ZHOU1,2, JIAN‑XIONG ZENG1,2, YU‑FEI DU1,2, FENG‑ZE SUN1,2, JIE‑YING YANG1,2, JUN‑YI HE1,2, YUE HUANG1,2, JIAN‑CHUAN XIA1,2 and DE‑SHENG WENG1,2 1State Key Laboratory of Oncology in South China, Collaborative Innovation Centre for Cancer Medicine; 2Department of Biotherapy, Sun Yat‑Sen University Cancer Center; 3Department of Radiation Oncology, Sun Yat‑Sen University Cancer Center, Guangzhou, Guangdong 510060, P.R. China Received July 31, 2020; Accepted December 18, 2020 DOI: 10.3892/ol.2021.12466 Abstract. Bladder cancer (BC) is the ninth most common immunohistochemistry and western blotting. In summary, lethal malignancy worldwide. Great efforts have been devoted KIF11 was significantly upregulated in BC and might act as to clarify the pathogenesis of BC, but the underlying molecular a potential prognostic biomarker. The present identification mechanisms remain unclear. To screen for the genes associated of DEGs and hub genes in BC may provide novel insight for with the progression and carcinogenesis of BC, three datasets investigating the molecular mechanisms of BC. were obtained from the Gene Expression Omnibus. A total of 37 tumor and 16 non‑cancerous samples were analyzed to Introduction identify differentially expressed genes (DEGs). Subsequently, 141 genes were identified, including 55 upregulated and Bladder cancer (BC) is the ninth most common malignancy 86 downregulated genes. The protein‑protein interaction worldwide with substantial morbidity and mortality. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Single-Cell RNA Sequencing Analysis of Human Neural Grafts Revealed Unexpected Cell Type Underlying the Genetic Risk of Parkinson’S Disease
Single-cell RNA Sequencing Analysis of Human Neural Grafts Revealed Unexpected Cell Type Underlying the Genetic Risk of Parkinson’s Disease Yingshan Wang1, Gang Wu2 1 Episcopal High School, 1200 N Quaker Ln, Alexandria, VA, USA, 22302 2 Fujian Sanbo Funeng Brain Hospital; Sanbo Brain Hospital Capital Medical University Abstract Parkinson’s disease (PD) is the second most common neurodegenerative disorder, affecting more than 6 million patients globally. Though previous studies have proposed several disease-related molecular pathways, how cell-type specific mechanisms contribute to the pathogenesis of PD is still mostly unknown. In this study, we analyzed single-cell RNA sequencing data of human neural grafts transplanted to the midbrains of rat PD models. Specifically, we performed cell-type identification, risk gene screening, and co-expression analysis. Our results revealed the unexpected genetic risk of oligodendrocytes as well as important pathways and transcription factors in PD pathology. The study may provide an overarching framework for understanding the cell non- autonomous effects in PD, inspiring new research hypotheses and therapeutic strategies. Keywords Parkinson’s Disease; Single-cell RNA Sequencing; Oligodendrocytes; Cell Non-autonomous; Co- expression Analysis; Transcription Factors 1 Table of Contents 1. Introduction ................................................................................................................................. 3 2. Methods...................................................................................................................................... -

Mitoxplorer, a Visual Data Mining Platform To
mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations Annie Yim, Prasanna Koti, Adrien Bonnard, Fabio Marchiano, Milena Dürrbaum, Cecilia Garcia-Perez, José Villaveces, Salma Gamal, Giovanni Cardone, Fabiana Perocchi, et al. To cite this version: Annie Yim, Prasanna Koti, Adrien Bonnard, Fabio Marchiano, Milena Dürrbaum, et al.. mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dy- namics and mutations. Nucleic Acids Research, Oxford University Press, 2020, 10.1093/nar/gkz1128. hal-02394433 HAL Id: hal-02394433 https://hal-amu.archives-ouvertes.fr/hal-02394433 Submitted on 4 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Nucleic Acids Research, 2019 1 doi: 10.1093/nar/gkz1128 Downloaded from https://academic.oup.com/nar/advance-article-abstract/doi/10.1093/nar/gkz1128/5651332 by Bibliothèque de l'université la Méditerranée user on 04 December 2019 mitoXplorer, a visual data mining platform to systematically analyze and visualize mitochondrial expression dynamics and mutations Annie Yim1,†, Prasanna Koti1,†, Adrien Bonnard2, Fabio Marchiano3, Milena Durrbaum¨ 1, Cecilia Garcia-Perez4, Jose Villaveces1, Salma Gamal1, Giovanni Cardone1, Fabiana Perocchi4, Zuzana Storchova1,5 and Bianca H. -
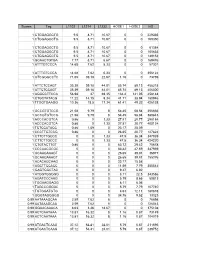
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108